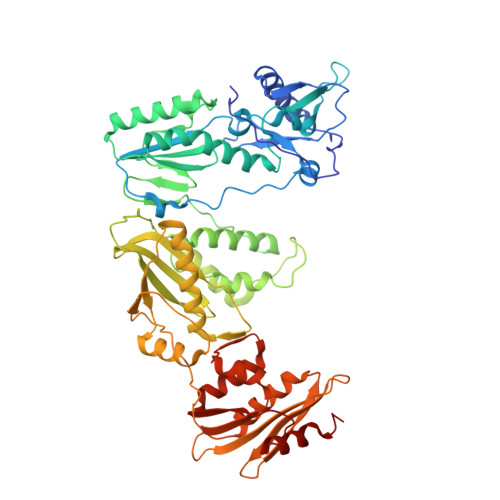
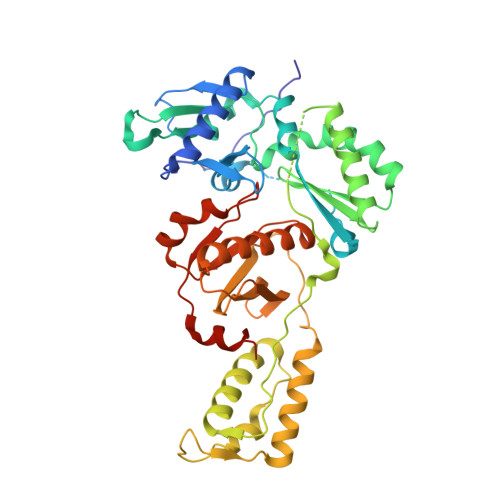
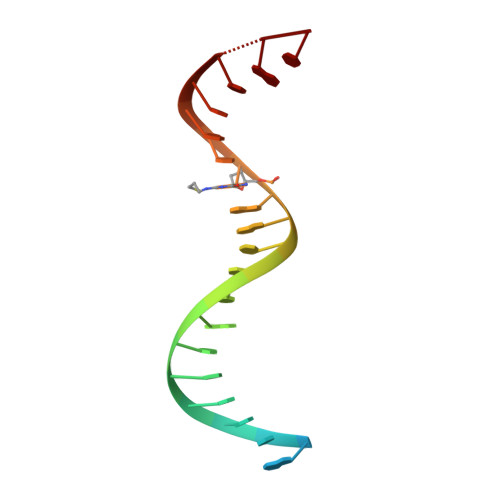
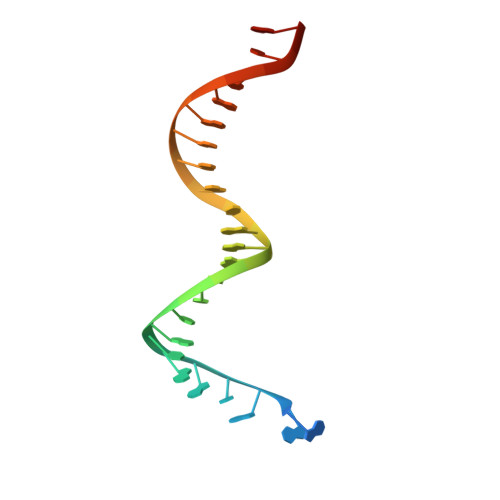
Structural basis of HIV inhibition by translocation-defective RT inhibitor 4'-ethynyl-2-fluoro-2'-deoxyadenosine (EFdA).
Salie, Z.L., Kirby, K.A., Michailidis, E., Marchand, B., Singh, K., Rohan, L.C., Kodama, E.N., Mitsuya, H., Parniak, M.A., Sarafianos, S.G.(2016) Proc Natl Acad Sci U S A 113: 9274-9279
- PubMed: 27489345
- DOI: https://doi.org/10.1073/pnas.1605223113
- Primary Citation of Related Structures:
5J2M, 5J2N, 5J2P, 5J2Q - PubMed Abstract:
4'-Ethynyl-2-fluoro-2'-deoxyadenosine (EFdA) is the most potent nucleoside analog inhibitor of HIV reverse transcriptase (RT). It retains a 3'-OH yet acts as a chain-terminating agent by diminishing translocation from the pretranslocation nucleotide-binding site (N site) to the posttranslocation primer-binding site (P site). Also, facile misincorporation of EFdA-monophosphate (MP) results in difficult-to-extend mismatched primers. To understand the high potency and unusual inhibition mechanism of EFdA, we solved RT crystal structures (resolutions from 2.4 to 2.9 Å) that include inhibition intermediates (i) before inhibitor incorporation (catalytic complex, RT/DNA/EFdA-triphosphate), (ii) after incorporation of EFdA-MP followed by dT-MP (RT/DNAEFdA-MP(P)• dT-MP(N) ), or (iii) after incorporation of two EFdA-MPs (RT/DNAEFdA-MP(P)• EFdA-MP(N) ); (iv) the latter was also solved with EFdA-MP mismatched at the N site (RT/DNAEFdA-MP(P)• EFdA-MP(*N) ). We report that the inhibition mechanism and potency of EFdA stem from interactions of its 4'-ethynyl at a previously unexploited conserved hydrophobic pocket in the polymerase active site. The high resolution of the catalytic complex structure revealed a network of ordered water molecules at the polymerase active site that stabilize enzyme interactions with nucleotide and DNA substrates. Finally, decreased translocation results from favorable interactions of primer-terminating EFdA-MP at the pretranslocation site and unfavorable posttranslocation interactions that lead to observed localized primer distortions.
- C. S. Bond Life Sciences Center, Department of Biochemistry, University of Missouri, Columbia, MO 65211;
Organizational Affiliation: