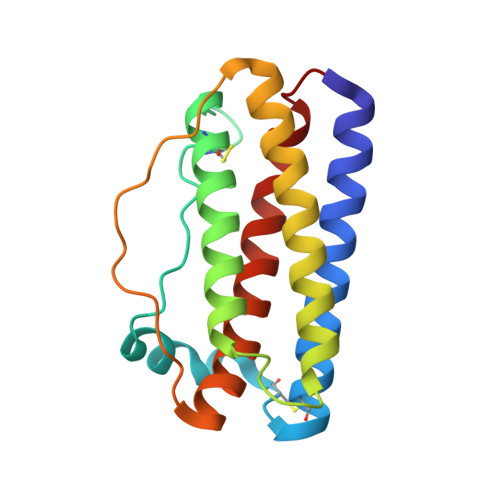
Structural insights into the backbone-circularized granulocyte colony-stimulating factor containing a short connector.
Miyafusa, T., Shibuya, R., Honda, S.(2018) Biochem Biophys Res Commun 500: 224-228
- PubMed: 29634929
- DOI: https://doi.org/10.1016/j.bbrc.2018.04.045
- Primary Citation of Related Structures:
5GW9 - PubMed Abstract:
Backbone circularization is a powerful approach for enhancing the structural stability of polypeptides. Herein, we present the crystal structure of the circularized variant of the granulocyte colony-stimulating factor (G-CSF) in which the terminal helical region was circularized using a short, two-amino acid connector. The structure revealed that the N- and C-termini were indeed connected by a peptide bond. The local structure of the C-terminal region transited from an α helix to 3 10 helix with a bend close to the N-terminal region, indicating that the structural change offset the insufficient length of the connector. This is the first-ever report of a crystal structure of the backbone of a circularized protein. It will facilitate the development of backbone circularization methodology.
- Biomedical Research Institute, The National Institute of Advanced Industrial Science and Technology, Central 6, 1-1-1 Higashi, Tsukuba, Ibaraki 305-8566, Japan.
Organizational Affiliation: