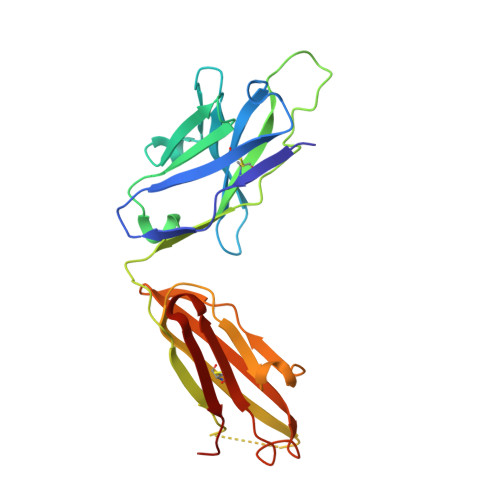
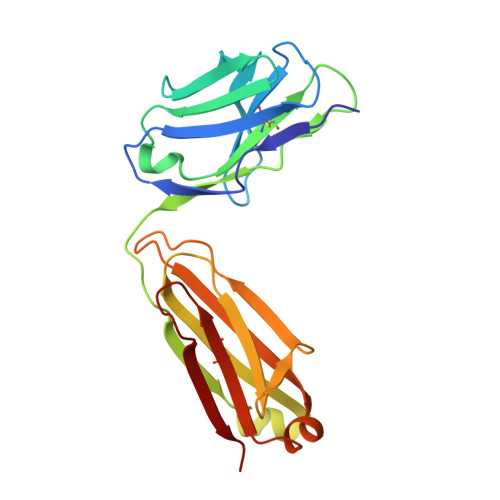
Molecular Basis for Antibody-Mediated Neutralization of New World Hemorrhagic Fever Mammarenaviruses.
Mahmutovic, S., Clark, L., Levis, S.C., Briggiler, A.M., Enria, D.A., Harrison, S.C., Abraham, J.(2015) Cell Host Microbe 18: 705-713
- PubMed: 26651946
- DOI: https://doi.org/10.1016/j.chom.2015.11.005
- Primary Citation of Related Structures:
5EN2 - PubMed Abstract:
In the Western hemisphere, at least five mammarenaviruses cause human viral hemorrhagic fevers with high case fatality rates. Junín virus (JUNV) is the only hemorrhagic fever virus for which transfusion of survivor immune plasma that contains neutralizing antibodies ("passive immunity") is an established treatment. Here, we report the structure of the JUNV surface glycoprotein receptor-binding subunit (GP1) bound to a neutralizing monoclonal antibody. The antibody engages the GP1 site that binds transferrin receptor 1 (TfR1)-the host cell surface receptor for all New World hemorrhagic fever mammarenaviruses-and mimics an important receptor contact. We show that survivor immune plasma contains antibodies that bind the same epitope. We propose that viral receptor-binding site accessibility explains the success of passive immunity against JUNV and that this functionally conserved epitope is a potential target for therapeutics and vaccines to limit infection by all New World hemorrhagic fever mammarenaviruses.
- Laboratory of Molecular Medicine, Boston Children's Hospital, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: