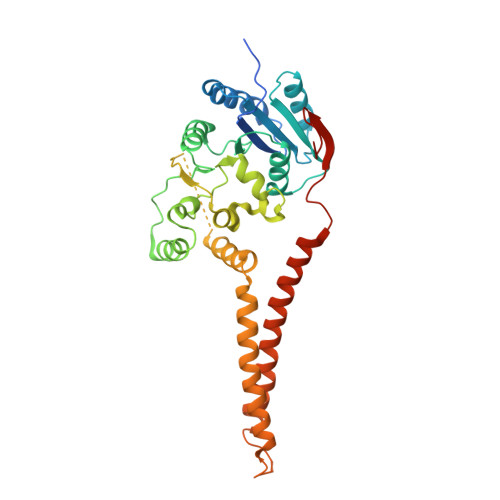
Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis.
Ardiccioni, C., Clarke, O.B., Tomasek, D., Issa, H.A., von Alpen, D.C., Pond, H.L., Banerjee, S., Rajashankar, K.R., Liu, Q., Guan, Z., Li, C., Kloss, B., Bruni, R., Kloppmann, E., Rost, B., Manzini, M.C., Shapiro, L., Mancia, F.(2016) Nat Commun 7: 10175-10175
- PubMed: 26729507
- DOI: https://doi.org/10.1038/ncomms10175
- Primary Citation of Related Structures:
5EKE, 5EKP - PubMed Abstract:
The attachment of a sugar to a hydrophobic polyisoprenyl carrier is the first step for all extracellular glycosylation processes. The enzymes that perform these reactions, polyisoprenyl-glycosyltransferases (PI-GTs) include dolichol phosphate mannose synthase (DPMS), which generates the mannose donor for glycosylation in the endoplasmic reticulum. Here we report the 3.0 Å resolution crystal structure of GtrB, a glucose-specific PI-GT from Synechocystis, showing a tetramer in which each protomer contributes two helices to a membrane-spanning bundle. The active site is 15 Å from the membrane, raising the question of how water-soluble and membrane-embedded substrates are brought into apposition for catalysis. A conserved juxtamembrane domain harbours disease mutations, which compromised activity in GtrB in vitro and in human DPM1 tested in zebrafish. We hypothesize a role of this domain in shielding the polyisoprenyl-phosphate for transport to the active site. Our results reveal the basis of PI-GT function, and provide a potential molecular explanation for DPM1-related disease.
- Department of Physiology and Cellular Biophysics, Columbia University, New York, New York 10032, USA.
Organizational Affiliation: