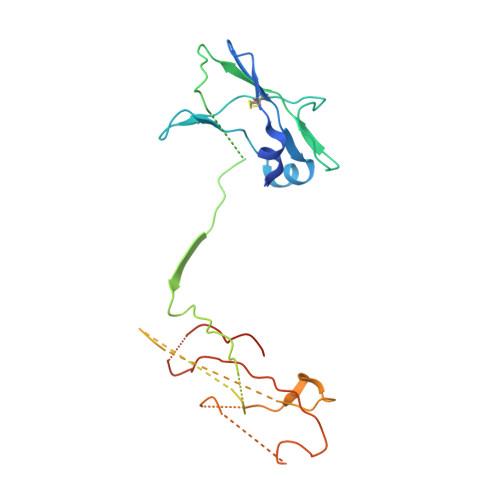
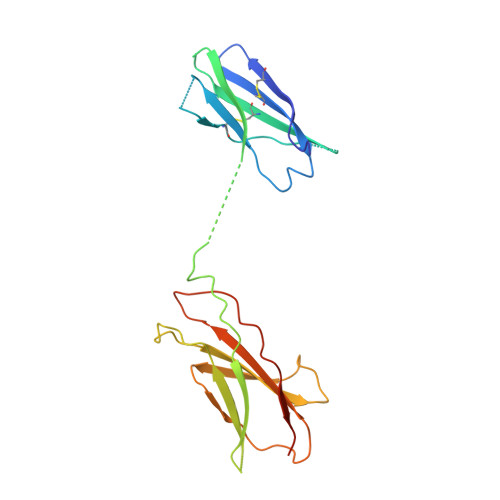
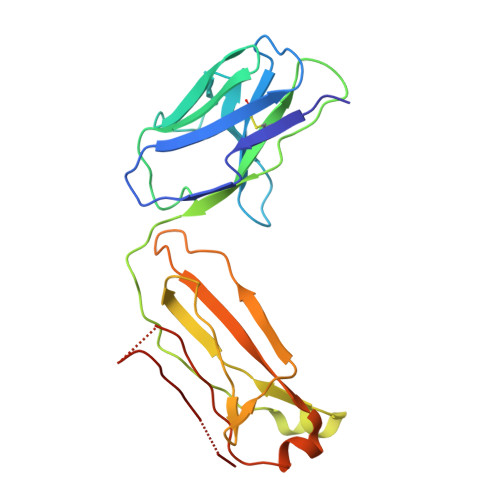
CSL311, a novel, potent, therapeutic monoclonal antibody for the treatment of diseases mediated by the common beta chain of the IL-3, GM-CSF and IL-5 receptors.
Panousis, C., Dhagat, U., Edwards, K.M., Rayzman, V., Hardy, M.P., Braley, H., Gauvreau, G.M., Hercus, T.R., Smith, S., Sehmi, R., McMillan, L., Dottore, M., McClure, B.J., Fabri, L.J., Vairo, G., Lopez, A.F., Parker, M.W., Nash, A.D., Wilson, N.J., Wilson, M.J., Owczarek, C.M.(2016) MAbs 8: 436-453
- PubMed: 26651396
- DOI: https://doi.org/10.1080/19420862.2015.1119352
- Primary Citation of Related Structures:
5DWU - PubMed Abstract:
The β common-signaling cytokines interleukin (IL)-3, granulocyte-macrophage colony stimulating factor (GM-CSF) and IL-5 stimulate pro-inflammatory activities of haematopoietic cells via a receptor complex incorporating cytokine-specific α and shared β common (βc, CD131) receptor. Evidence from animal models and recent clinical trials demonstrate that these cytokines are critical mediators of the pathogenesis of inflammatory airway disease such as asthma. However, no therapeutic agents, other than steroids, that specifically and effectively target inflammation mediated by all 3 of these cytokines exist. We employed phage display technology to identify and optimize a novel, human monoclonal antibody (CSL311) that binds to a unique epitope that is specific to the cytokine-binding site of the human βc receptor. The binding epitope of CSL311 on the βc receptor was defined by X-ray crystallography and site-directed mutagenesis. CSL311 has picomolar binding affinity for the human βc receptor, and at therapeutic concentrations is a highly potent antagonist of the combined activities of IL-3, GM-CSF and IL-5 on primary eosinophil survival in vitro. Importantly, CSL311 inhibited the survival of inflammatory cells present in induced sputum from human allergic asthmatic subjects undergoing allergen bronchoprovocation. Due to its high potency and ability to simultaneously suppress the activity of all 3 β common cytokines, CSL311 may provide a new strategy for the treatment of chronic inflammatory diseases where the human βc receptor is central to pathogenesis. The coordinates for the βc/CSL311 Fab complex structure have been deposited with the RCSB Protein Data Bank (PDB 5DWU).
- a Research and Development, CSL Limited; Bio21 Molecular Science and Biotechnology Institute , Parkville Victoria , 3010 , Australia.
Organizational Affiliation: