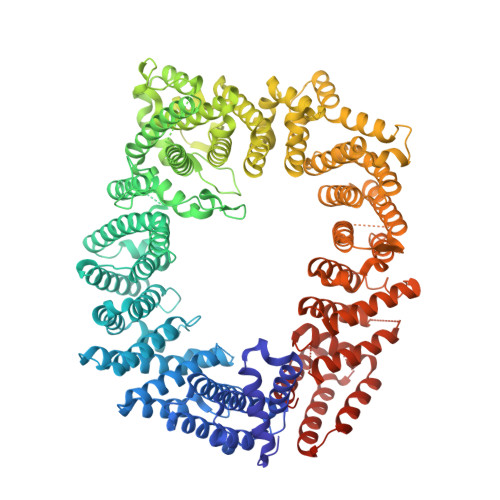
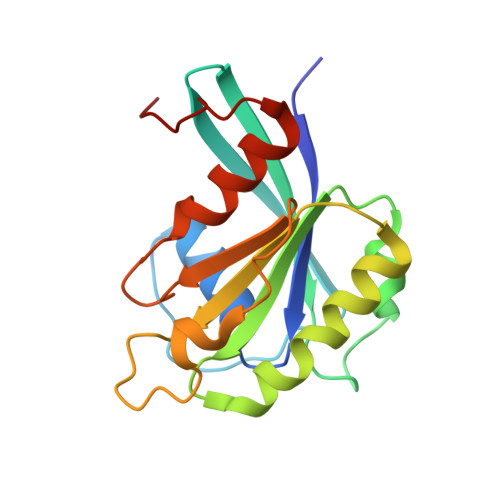
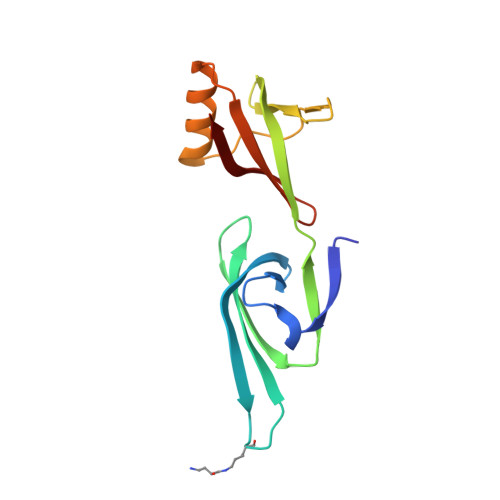
Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A.
Aksu, M., Trakhanov, S., Gorlich, D.(2016) Nat Commun 7: 11952-11952
- PubMed: 27306458
- DOI: https://doi.org/10.1038/ncomms11952
- Primary Citation of Related Structures:
5DLQ - PubMed Abstract:
Xpo4 is a bidirectional nuclear transport receptor that mediates nuclear export of eIF5A and Smad3 as well as import of Sox2 and SRY. How Xpo4 recognizes such a variety of cargoes is as yet unknown. Here we present the crystal structure of the RanGTP·Xpo4·eIF5A export complex at 3.2 Å resolution. Xpo4 has a similar structure as CRM1, but the NES-binding site is occluded, and a new interaction site evolved that recognizes both globular domains of eIF5A. eIF5A contains hypusine, a unique amino acid with two positive charges, which is essential for cell viability and eIF5A function in translation. The hypusine docks into a deep, acidic pocket of Xpo4 and is thus a critical element of eIF5A's complex export signature. This further suggests that Xpo4 recognizes other cargoes differently, and illustrates how Xpo4 suppresses - in a chaperone-like manner - undesired interactions of eIF5A inside nuclei.
- Department of Cellular Logistics, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany.
Organizational Affiliation: