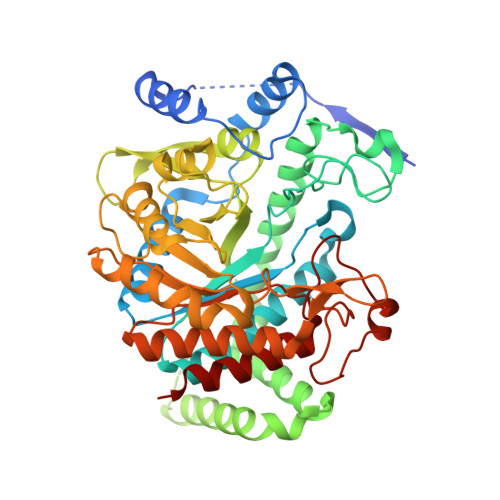
Remote Control by Inter-Enzyme Allostery: A Novel Paradigm for Regulation of the Shikimate Pathway.
Munack, S., Roderer, K., Okvist, M., Kamarauskaite, J., Sasso, S., van Eerde, A., Kast, P., Krengel, U.(2016) J Mol Biology 428: 1237-1255
- PubMed: 26776476
- DOI: https://doi.org/10.1016/j.jmb.2016.01.001
- Primary Citation of Related Structures:
5CKV, 5CKX - PubMed Abstract:
DAHP synthase and chorismate mutase catalyze key steps in the shikimate biosynthetic pathway en route to aromatic amino acids. In Mycobacterium tuberculosis, chorismate mutase (MtCM; Rv0948c), located at the branch point toward phenylalanine and tyrosine, has poor activity on its own. However, it is efficiently activated by the first enzyme of the pathway, DAHP synthase (MtDS; Rv2178c), through formation of a non-covalent MtCM-MtDS complex. Here, we show how MtDS serves as an allosteric platform for feedback regulation of both enzymes, using X-ray crystallography, small-angle X-ray scattering, size-exclusion chromatography, and multi-angle light scattering. Crystal structures of the fully inhibited MtDS and the allosterically down-regulated MtCM-MtDS complex, solved at 2.8 and 2.7Å, respectively, reveal how effector binding at the internal MtDS subunit interfaces regulates the activity of MtDS and MtCM. While binding of all three metabolic end products to MtDS shuts down the entire pathway, the binding of phenylalanine jointly with tyrosine releases MtCM from the MtCM-MtDS complex, hence suppressing MtCM activation by 'inter-enzyme allostery'. This elegant regulatory principle, invoking a transient allosteric enzyme interaction, seems to be driven by dynamics and is likely a general strategy used by nature.
- Department of Chemistry, University of Oslo, NO-0315 Oslo, Norway.
Organizational Affiliation: