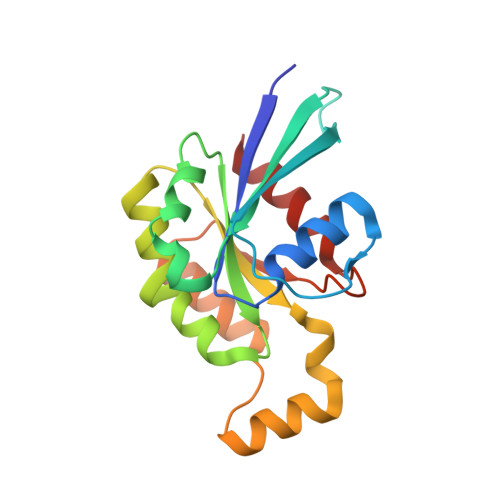
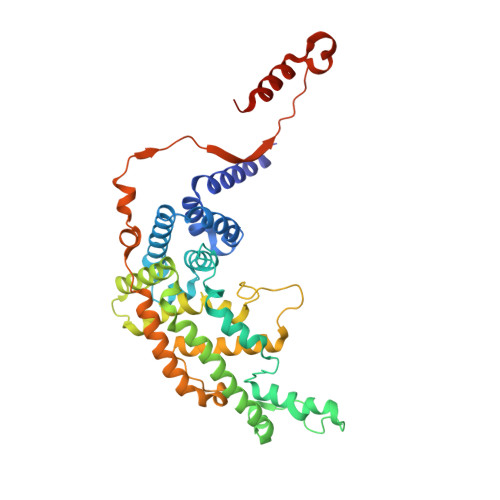
The Structural Basis for Cdc42-Induced Dimerization of IQGAPs.
LeCour, L., Boyapati, V.K., Liu, J., Li, Z., Sacks, D.B., Worthylake, D.K.(2016) Structure 24: 1499-1508
- PubMed: 27524202
- DOI: https://doi.org/10.1016/j.str.2016.06.016
- Primary Citation of Related Structures:
5CJP - PubMed Abstract:
In signaling, Rho-family GTPases bind effector proteins and alter their behavior. Here we present the crystal structure of Cdc42·GTP bound to the GTPase-activating protein (GAP)-related domain (GRD) of IQGAP2. Four molecules of Cdc42 are bound to two GRD molecules, which bind each other in a parallel dimer. Two Cdc42s bind very similarly to the Ras/RasGAP interaction, while the other two bind primarily to "extra domain" sequences from both GRDs, tying the GRDs together. Calorimetry confirms two-site binding of Cdc42·GTP for the GRDs of both IQGAP2 and IQGAP1. Mutation of important extra domain residues reduces binding to single-site and abrogates Cdc42 binding to a much larger IQGAP1 fragment. Importantly, Rac1·GTP displays only single-site binding to the GRDs, indicating that only Cdc42 promotes IQGAP dimerization. The structure identifies an unexpected role for Cdc42 in protein dimerization, thus expanding the repertoire of interactions of Ras family proteins with their targets.
- Department of Biochemistry and Molecular Biology, Louisiana State University Health Sciences Center, New Orleans, LA 70112, USA.
Organizational Affiliation: