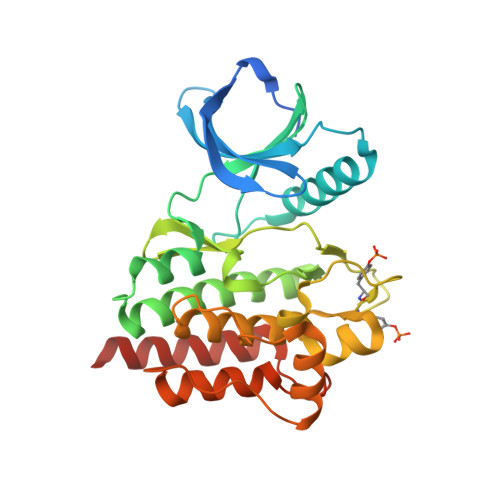
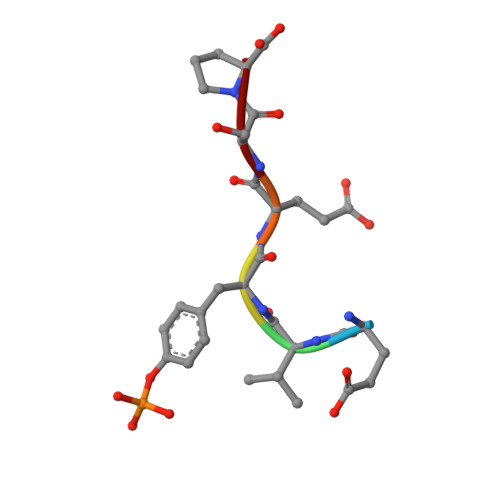
Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP).
Lovering, F., Aevazelis, C., Chang, J., Dehnhardt, C., Fitz, L., Han, S., Janz, K., Lee, J., Kaila, N., McDonald, J., Moore, W., Moretto, A., Papaioannou, N., Richard, D., Ryan, M.S., Wan, Z.K., Thorarensen, A.(2016) ChemMedChem 11: 217-233
- PubMed: 26381330
- DOI: https://doi.org/10.1002/cmdc.201500333
- Primary Citation of Related Structures:
5C26, 5C27 - PubMed Abstract:
There has been significant interest in spleen tyrosine kinase (Syk) owing to its role in a number of disease states, including autoimmunity, inflammation, and cancer. Ongoing therapeutic programs have resulted in several compounds that are now in clinical use. Herein we report our optimization of the imidazopyrazine core scaffold of Syk inhibitors through the use of empirical and computational approaches. Free-energy perturbation (FEP) methods with MCPRO+ were undertaken to calculate the relative binding free energies for several alternate scaffolds. FEP was first applied retrospectively to determine if there is any predictive value; this resulted in 12 of 13 transformations being predicted in a directionally correct manner. FEP was then applied in a prospective manner to evaluate 17 potential targets, resulting in the realization of imidazotriazine 17 (3-(4-(3,4-dimethoxyphenylamino)imidazo[1,2-f][1,2,4]triazin-2-yl)benzamide), which shows a tenfold improvement in activity relative to the parent compound and no increase in atom count. Optimization of 17 led to compounds with nanomolar cellular activity.
- Worldwide Medicinal Chemistry, Pfizer Worldwide R&D, 610 Main Street, Cambridge, MA, 02139, USA. frank.lovering@pfizer.com.
Organizational Affiliation: