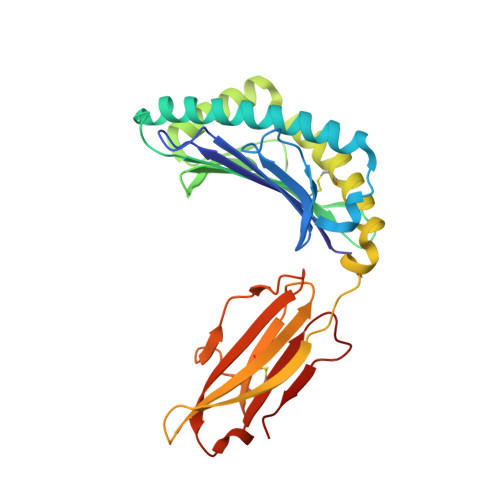
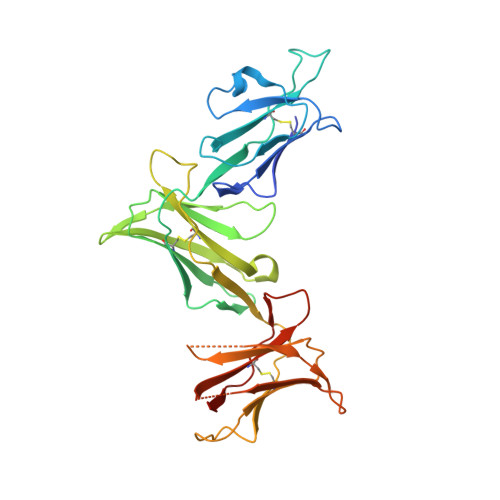
Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition
Saunders, P.M., Pymm, P., Pietra, G., Hughes, V.A., Hitchen, C., O'Connor, G.M., Loiacono, F., Widjaja, J., Price, D.A., Falco, M., Mingari, M.C., Moretta, L., McVicar, D.W., Rossjohn, J., Brooks, A.G., Vivian, J.P.(2016) J Exp Medicine 213: 791-807
- PubMed: 27045007
- DOI: https://doi.org/10.1084/jem.20152023
- Primary Citation of Related Structures:
5B38, 5B39 - PubMed Abstract:
Natural killer (NK) cells play a key role in immunity, but how HLA class I (HLA-I) and killer cell immunoglobulin-like receptor 3DL1 (KIR3DL1) polymorphism impacts disease outcome remains unclear. KIR3DL1 (*001/*005/*015) tetramers were screened for reactivity against a panel of HLA-I molecules. This revealed different and distinct hierarchies of specificity for each KIR3DL1 allotype, with KIR3DL1*005 recognizing the widest array of HLA-I ligands. These differences were further reflected in functional studies using NK clones expressing these specific KIR3DL1 allotypes. Unexpectedly, the Ile/Thr80 dimorphism in the Bw4-motif did not categorically define strong/weak KIR3DL1 recognition. Although the KIR3DL1*001, *005, and *015 polymorphisms are remote from the KIR3DL1-HLA-I interface, the structures of these three KIR3DL1-HLA-I complexes showed that the broader HLA-I specificity of KIR3DL1*005 correlated with an altered KIR3DL1*005 interdomain positioning and increased mobility within its ligand-binding site. Collectively, we provide a generic framework for understanding the impact of KIR3DL1 polymorphism on the recognition of HLA-I allomorphs.
- Department of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity, The University of Melbourne, Parkville, Victoria 3010, Australia.
Organizational Affiliation: