Genetically encoded phenyl azide photochemistry drives positive and negative functional modulation of a red fluorescent protein
Reddington, S.C., Driezis, S., Hartley, A.M., Watson, P.D., Rizkallah, P.J., Jones, D.D.(2015) RSC Adv 5: 77734-77738
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2015) RSC Adv 5: 77734-77738
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MCherry fluorescent protein | 217 | Anaplasma marginale | Mutation(s): 1 Gene Names: mCherry | 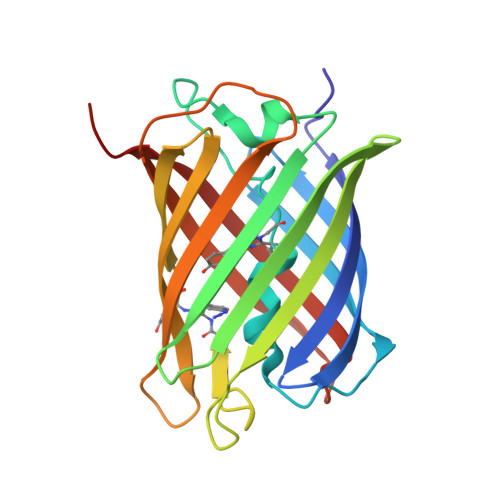 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | D [auth A] E [auth A] F [auth B] G [auth B] H [auth C] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| EDO Query on EDO | J [auth C] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Modified Residues 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| 4II Query on 4II | A, B, C | L-PEPTIDE LINKING | C9 H10 N4 O2 |  | PHE |
| CH6 Query on CH6 | A, B, C | L-PEPTIDE LINKING | C16 H19 N3 O4 S |  | MET, TYR, GLY |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 43.28 | α = 90 |
| b = 103.43 | β = 90 |
| c = 150.72 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Aimless | data scaling |
| PHASER | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| xia2 | data reduction |
| PHASER | phasing |