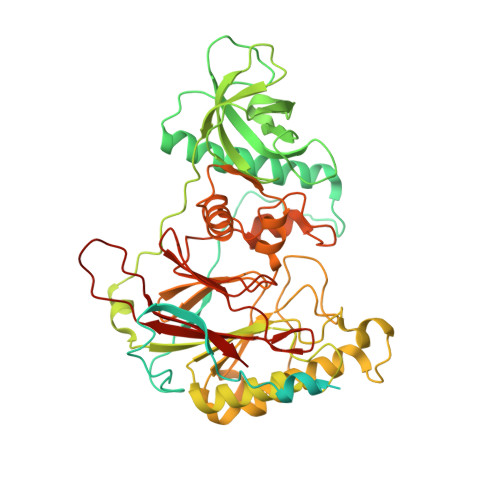
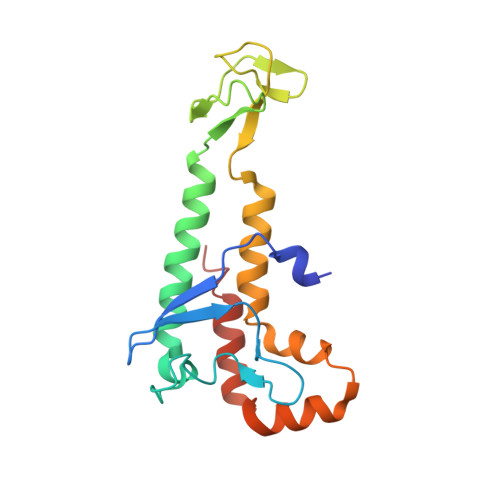
Crystal Structure of the Human Pol alpha B Subunit in Complex with the C-terminal Domain of the Catalytic Subunit.
Suwa, Y., Gu, J., Baranovskiy, A.G., Babayeva, N.D., Pavlov, Y.I., Tahirov, T.H.(2015) J Biological Chem 290: 14328-14337
- PubMed: 25847248
- DOI: https://doi.org/10.1074/jbc.M115.649954
- Primary Citation of Related Structures:
4Y97 - PubMed Abstract:
In eukaryotic DNA replication, short RNA-DNA hybrid primers synthesized by primase-DNA polymerase α (Prim-Pol α) are needed to start DNA replication by the replicative DNA polymerases, Pol δ and Pol ϵ. The C terminus of the Pol α catalytic subunit (p180C) in complex with the B subunit (p70) regulates the RNA priming and DNA polymerizing activities of Prim-Pol α. It tethers Pol α and primase, facilitating RNA primer handover from primase to Pol α. To understand these regulatory mechanisms and to reveal the details of human Pol α organization, we determined the crystal structure of p70 in complex with p180C. The structured portion of p70 includes a phosphodiesterase (PDE) domain and an oligonucleotide/oligosaccharide binding (OB) domain. The N-terminal domain and the linker connecting it to the PDE domain are disordered in the reported crystal structure. The p180C adopts an elongated asymmetric saddle shape, with a three-helix bundle in the middle and zinc-binding modules (Zn1 and Zn2) on each side. The extensive p180C-p70 interactions involve 20 hydrogen bonds and a number of hydrophobic interactions resulting in an extended buried surface of 4080 Å(2). Importantly, in the structure of the p180C-p70 complex with full-length p70, the residues from the N-terminal to the OB domain contribute to interactions with p180C. The comparative structural analysis revealed both the conserved features and the differences between the human and yeast Pol α complexes.
- From the Eppley Institute for Research in Cancer and Allied Diseases and.
Organizational Affiliation: