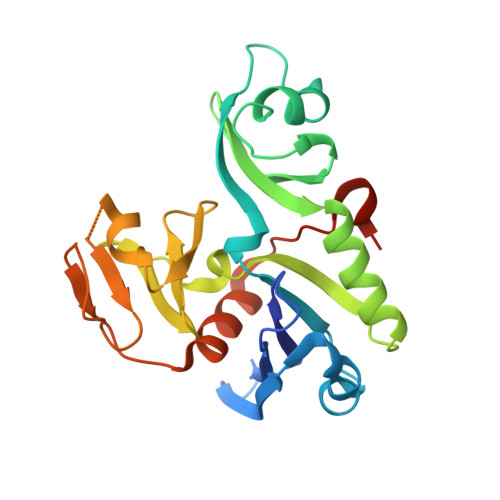
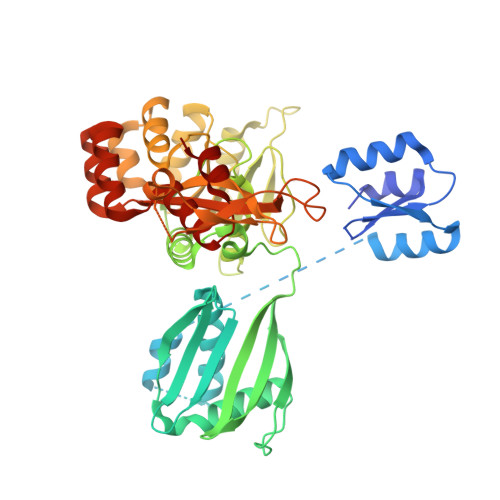
Crystal structure of the full-length ATPase GspE from the Vibrio vulnificus type II secretion system in complex with the cytoplasmic domain of GspL.
Lu, C., Korotkov, K.V., Hol, W.G.(2014) J Struct Biol 187: 223-235
- PubMed: 25092625
- DOI: https://doi.org/10.1016/j.jsb.2014.07.006
- Primary Citation of Related Structures:
4PHT - PubMed Abstract:
The type II secretion system (T2SS) is present in many Gram-negative bacteria and is responsible for secreting a large number of folded proteins, including major virulence factors, across the outer membrane. The T2SS consists of 11-15 different proteins most of which are present in multiple copies in the assembled secretion machinery. The ATPase GspE, essential for the functioning of the T2SS, contains three domains (N1E, N2E and CTE) of which the N1E domain is associated with the cytoplasmic domain of the inner membrane protein GspL. Here we describe and analyze the structure of the GspE•cyto-GspL complex from Vibrio vulnificus in the presence of an ATP analog, AMPPNP. There are three such ∼83 kDa complexes per asymmetric unit with essentially the same structure. The N2E and CTE domains of a single V. vulnificus GspE subunit adopt a mutual orientation that has not been seen before in any of the previous GspE structures, neither in structures of related ATPases from other secretion systems. This underlines the tremendous conformational flexibility of the T2SS secretion ATPase. Cyto-GspL interacts not only with the N1E domain, but also with the CTE domain and is even in contact with AMPPNP. Moreover, the cyto-GspL domains engage in two types of mutual interactions, resulting in two essentially identical, but crystallographically independent, "cyto-GspL rods" that run throughout the crystal. Very similar rods are present in previous crystals of cyto-GspL and of the N1E•cyto-GspL complex. This arrangement, now seen four times in three entirely different crystal forms, involves contacts between highly conserved residues suggesting a role in the biogenesis or the secretion mechanism or both of the T2SS.
- Department of Biochemistry and Biomolecular Structure Center, University of Washington, Seattle, WA 98195, United States.
Organizational Affiliation: