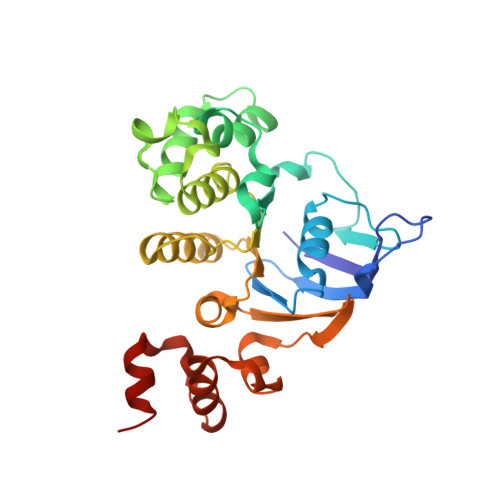
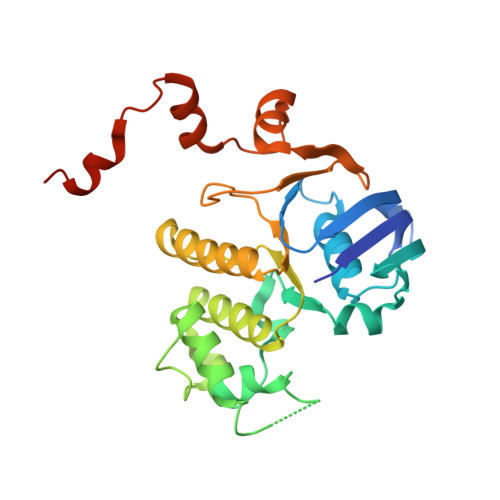
ATP binding drives substrate capture in an ECF transporter by a release-and-catch mechanism.
Karpowich, N.K., Song, J.M., Cocco, N., Wang, D.N.(2015) Nat Struct Mol Biol 22: 565-571
- PubMed: 26052893
- DOI: https://doi.org/10.1038/nsmb.3040
- Primary Citation of Related Structures:
4ZIR - PubMed Abstract:
ECF transporters are a family of active transporters for vitamins. They are composed of four subunits: a membrane-embedded substrate-binding subunit (EcfS), a transmembrane coupling subunit (EcfT) and two ATP-binding-cassette ATPases (EcfA and EcfA'). We have investigated the mechanism of the ECF transporter for riboflavin from the pathogen Listeria monocytogenes, LmECF-RibU. Using structural and biochemical approaches, we found that ATP binding to the EcfAA' ATPases drives a conformational change that dissociates the S subunit from the EcfAA'T ECF module. Upon release from the ECF module, the RibU S subunit then binds the riboflavin transport substrate. We also find that S subunits for distinct substrates compete for the ATP-bound state of the ECF module. Our results explain how ECF transporters capture the transport substrate and reproduce the in vivo observations on S-subunit competition for which the family was named.
- Skirball Institute of Biomolecular Medicine, New York University School of Medicine, New York, New York, USA.
Organizational Affiliation: