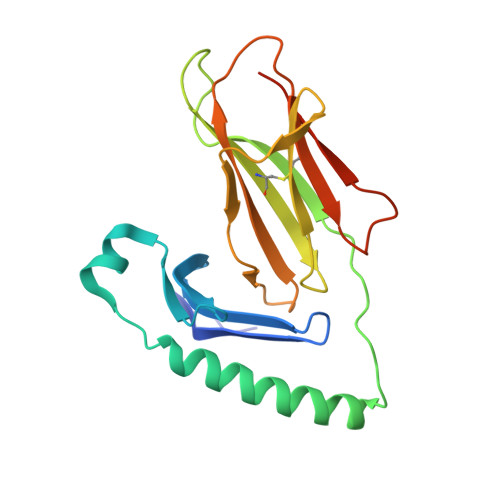
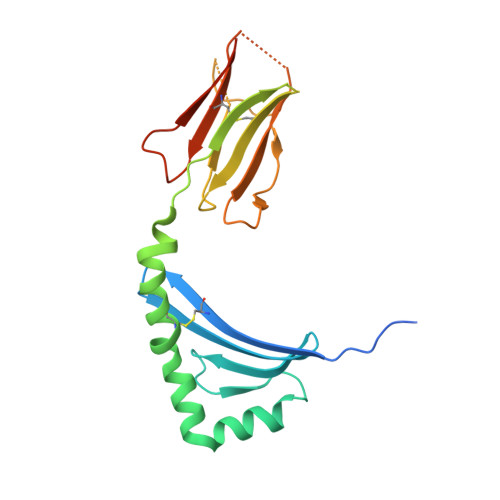
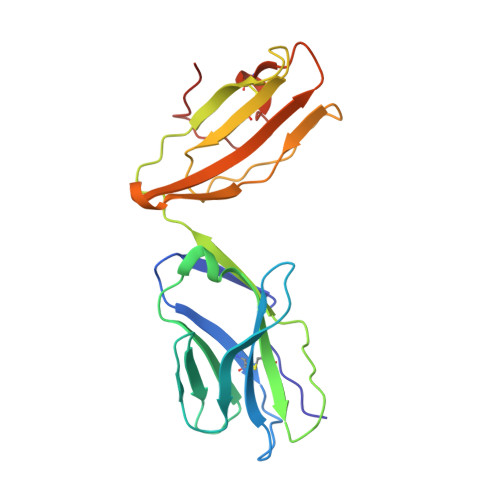
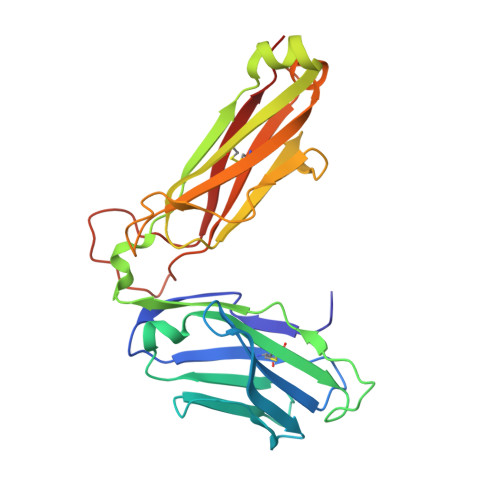
Determinants of Gliadin-Specific T Cell Selection in Celiac Disease.
Petersen, J., van Bergen, J., Loh, K.L., Kooy-Winkelaar, Y., Beringer, D.X., Thompson, A., Bakker, S.F., Mulder, C.J., Ladell, K., McLaren, J.E., Price, D.A., Rossjohn, J., Reid, H.H., Koning, F.(2015) J Immunol 194: 6112-6122
- PubMed: 25948817
- DOI: https://doi.org/10.4049/jimmunol.1500161
- Primary Citation of Related Structures:
4Z7U, 4Z7V, 4Z7W - PubMed Abstract:
In HLA-DQ8-associated celiac disease (CD), the pathogenic T cell response is directed toward an immunodominant α-gliadin-derived peptide (DQ8-glia-α1). However, our knowledge of TCR gene usage within the primary intestinal tissue of HLA-DQ8 (+) CD patients is limited. We identified two populations of HLA-DQ8-glia-α1 tetramer(+) CD4(+) T cells that were essentially undetectable in biopsy samples from patients on a gluten-free diet but expanded rapidly and specifically after antigenic stimulation. Distinguished by expression of TRBV9, both T cell populations displayed biased clonotypic repertoires and reacted similarly against HLA-DQ8-glia-α1. In particular, TRBV9 paired most often with TRAV26-2, whereas the majority of TRBV9(-) TCRs used TRBV6-1 with no clear TRAV gene preference. Strikingly, both tetramer(+)/TRBV9(+) and tetramer(+)/TRBV9(-) T cells possessed a non-germline-encoded arginine residue in their CDR3α and CDR3β loops, respectively. Comparison of the crystal structures of three TRBV9(+) TCRs and a TRBV9(-) TCR revealed that, as a result of distinct TCR docking modes, the HLA-DQ8-glia-α1 contacts mediated by the CDR3-encoded arginine were almost identical between TRBV9(+) and TRBV9(-) TCRs. In all cases, this interaction centered on two hydrogen bonds with a specific serine residue in the bound peptide. Replacement of serine with alanine at this position abrogated TRBV9(+) and TRBV9(-) clonal T cell proliferation in response to HLA-DQ8-glia-α1. Gluten-specific memory CD4(+) T cells with structurally and functionally conserved TCRs therefore predominate in the disease-affected tissue of patients with HLA-DQ8-mediated CD.
- Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria 3800, Australia; Australian Research Council Centre of Excellence in Advanced Molecular Imaging, Monash University, Clayton, Victoria 3800, Australia;
Organizational Affiliation: