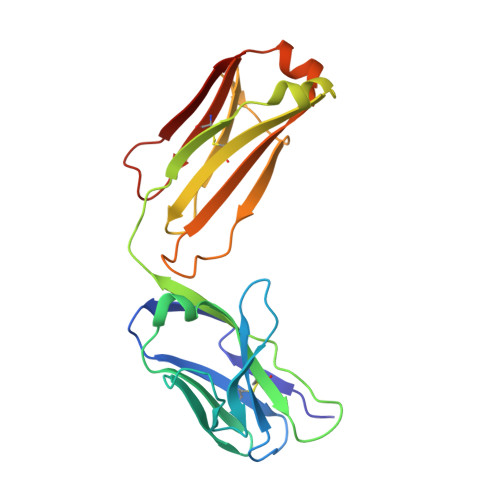
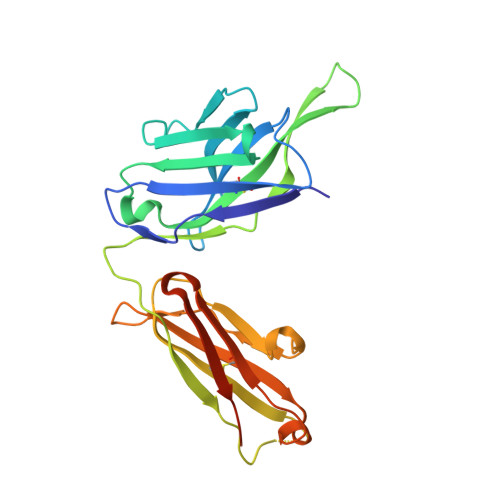
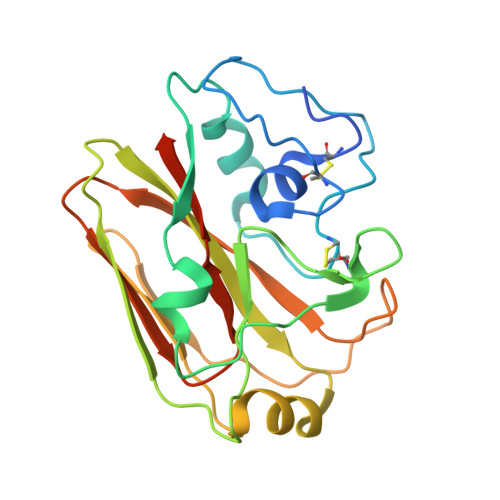
Viral receptor-binding site antibodies with diverse germline origins.
Schmidt, A.G., Therkelsen, M.D., Stewart, S., Kepler, T.B., Liao, H.X., Moody, M.A., Haynes, B.F., Harrison, S.C.(2015) Cell 161: 1026-1034
- PubMed: 25959776
- DOI: https://doi.org/10.1016/j.cell.2015.04.028
- Primary Citation of Related Structures:
4YJZ, 4YK4 - PubMed Abstract:
Vaccines for rapidly evolving pathogens will confer lasting immunity if they elicit antibodies recognizing conserved epitopes, such as a receptor-binding site (RBS). From characteristics of an influenza-virus RBS-directed antibody, we devised a signature motif to search for similar antibodies. We identified, from three vaccinees, over 100 candidates encoded by 11 different VH genes. Crystal structures show that antibodies in this class engage the hemagglutinin RBS and mimic binding of the receptor, sialic acid, by supplying a critical dipeptide on their projecting, heavy-chain third complementarity determining region. They share contacts with conserved, receptor-binding residues but contact different residues on the RBS periphery, limiting the likelihood of viral escape when several such antibodies are present. These data show that related modes of RBS recognition can arise from different germline origins and mature through diverse affinity maturation pathways. Immunogens focused on an RBS-directed response will thus have a broad range of B cell targets.
- Laboratory of Molecular Medicine, Children's Hospital, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: