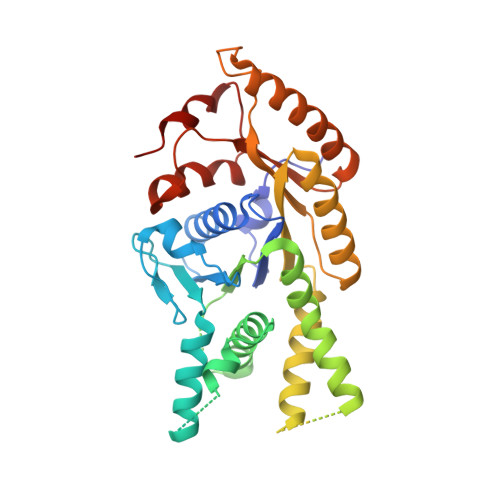
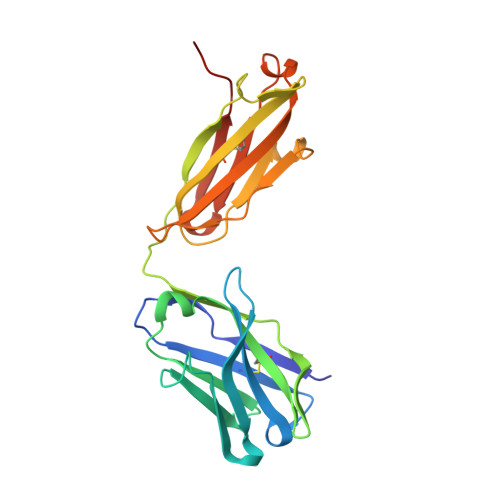
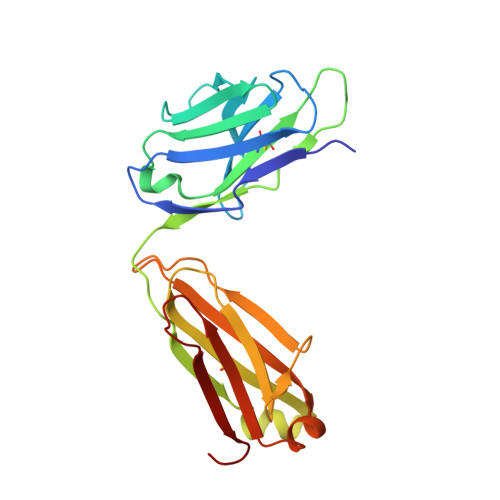
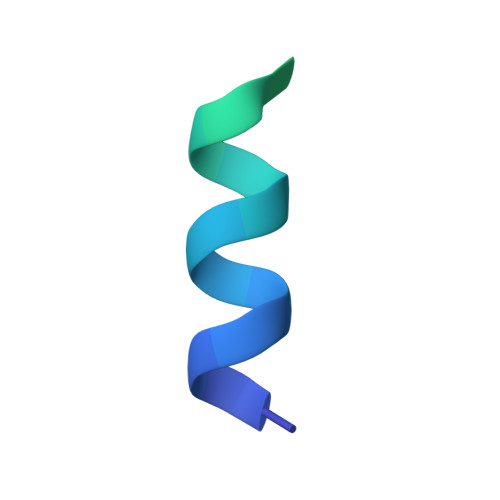
Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Mateja, A., Paduch, M., Chang, H.Y., Szydlowska, A., Kossiakoff, A.A., Hegde, R.S., Keenan, R.J.(2015) Science 347: 1152-1155
- PubMed: 25745174
- DOI: https://doi.org/10.1126/science.1261671
- Primary Citation of Related Structures:
4XTR, 4XVU, 4XWO - PubMed Abstract:
Tail-anchored (TA) proteins are a physiologically important class of membrane proteins targeted to the endoplasmic reticulum by the conserved guided-entry of TA proteins (GET) pathway. During transit, their hydrophobic transmembrane domains (TMDs) are chaperoned by the cytosolic targeting factor Get3, but the molecular nature of the functional Get3-TA protein targeting complex remains unknown. We reconstituted the physiologic assembly pathway for a functional targeting complex and showed that it comprises a TA protein bound to a Get3 homodimer. Crystal structures of Get3 bound to different TA proteins showed an α-helical TMD occupying a hydrophobic groove that spans the Get3 homodimer. Our data elucidate the mechanism of TA protein recognition and shielding by Get3 and suggest general principles of hydrophobic domain chaperoning by cellular targeting factors.
- Department of Biochemistry and Molecular Biology, The University of Chicago, 929 East 57th Street, Chicago, IL 60637, USA.
Organizational Affiliation: