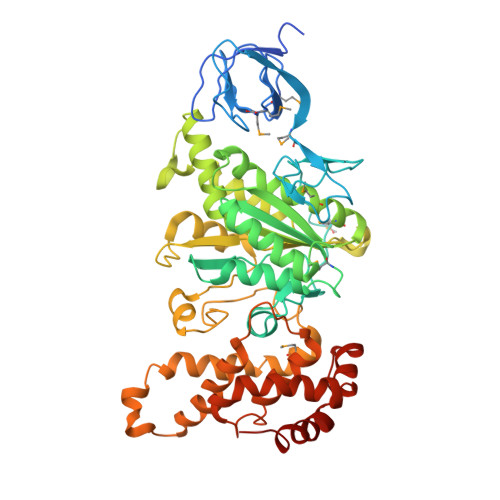
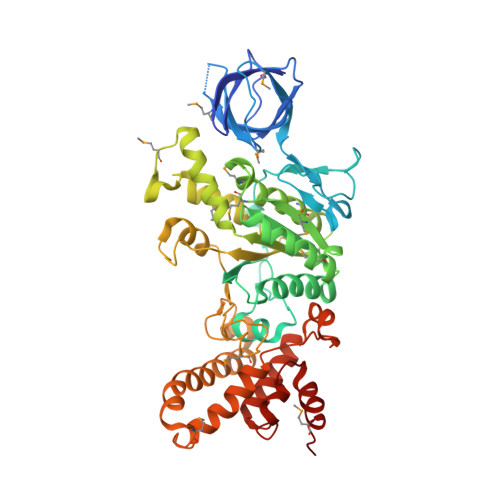
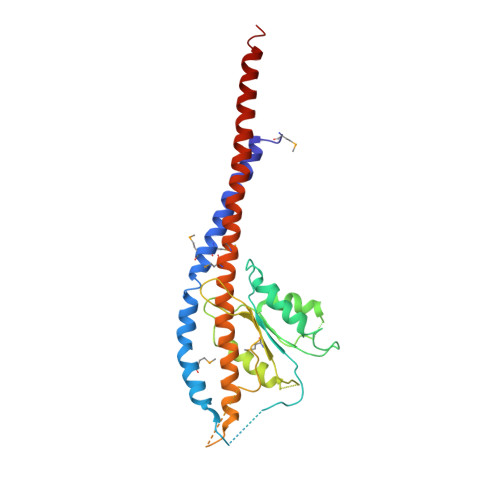
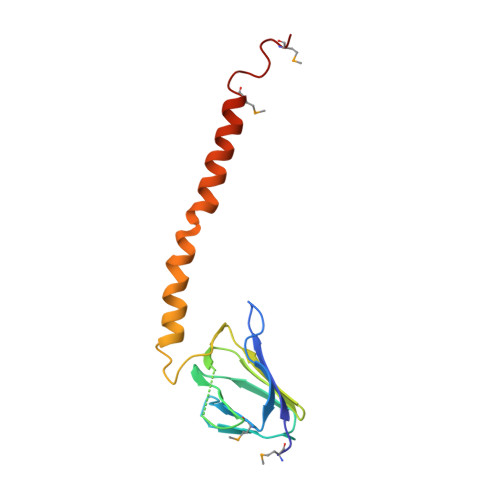
Structure of a thermophilic F1 -ATPase inhibited by an epsilon-subunit: deeper insight into the epsilon-inhibition mechanism.
Shirakihara, Y., Shiratori, A., Tanikawa, H., Nakasako, M., Yoshida, M., Suzuki, T.(2015) FEBS J 282: 2895-2913
- PubMed: 26032434
- DOI: https://doi.org/10.1111/febs.13329
- Primary Citation of Related Structures:
4XD7 - PubMed Abstract:
F1-ATPase (F1) is the catalytic sector in F(o)F1-ATP synthase that is responsible for ATP production in living cells. In catalysis, its three catalytic β-subunits undergo nucleotide occupancy-dependent and concerted open-close conformational changes that are accompanied by rotation of the γ-subunit. Bacterial and chloroplast F1 are inhibited by their own ε-subunit. In the ε-inhibited Escherichia coli F1 structure, the ε-subunit stabilizes the overall conformation (half-closed, closed, open) of the β-subunits by inserting its C-terminal helix into the α3β3 cavity. The structure of ε-inhibited thermophilic F1 is similar to that of E. coli F1, showing a similar conformation of the ε-subunit, but the thermophilic ε-subunit stabilizes another unique overall conformation (open, closed, open) of the β-subunits. The ε-C-terminal helix 2 and hook are conserved between the two structures in interactions with target residues and in their positions. Rest of the ε-C-terminal domains are in quite different conformations and positions, and have different modes of interaction with targets. This region is thought to serve ε-inhibition differently. For inhibition, the ε-subunit contacts the second catches of some of the β- and α-subunits, the N- and C-terminal helices, and some of the Rossmann fold segments. Those contacts, as a whole, lead to positioning of those β- and α- second catches in ε-inhibition-specific positions, and prevent rotation of the γ-subunit. Some of the structural features are observed even in IF1 inhibition in mitochondrial F1.
- National Institute of Genetics, Mishima, Japan.
Organizational Affiliation: