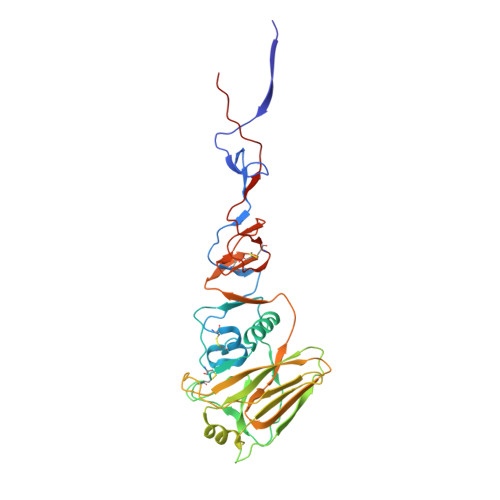
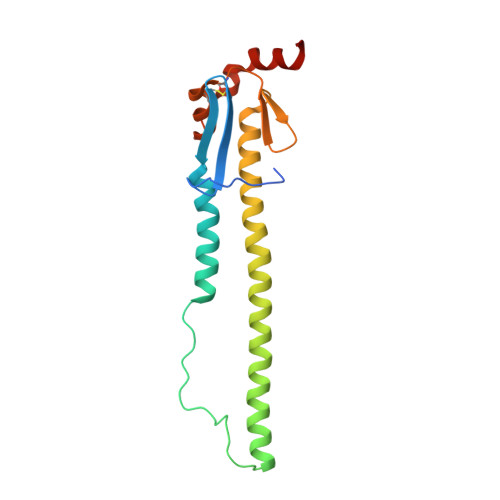
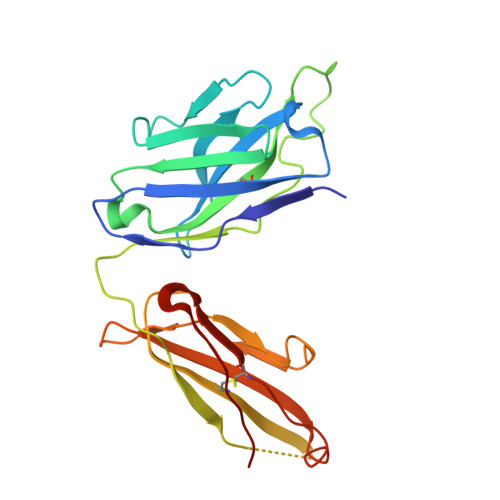
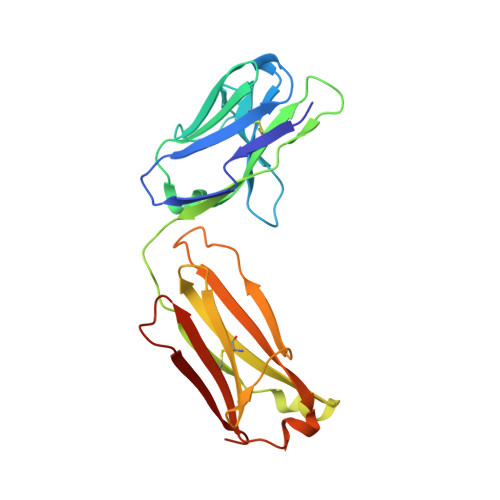
A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus.
Wu, Y., Cho, M., Shore, D., Song, M., Choi, J., Jiang, T., Deng, Y.Q., Bourgeois, M., Almli, L., Yang, H., Chen, L.M., Shi, Y., Qi, J., Li, A., Yi, K.S., Chang, M., Bae, J.S., Lee, H., Shin, J., Stevens, J., Hong, S., Qin, C.F., Gao, G.F., Chang, S.J., Donis, R.O.(2015) Nat Commun 6: 7708-7708
- PubMed: 26196962
- DOI: https://doi.org/10.1038/ncomms8708
- Primary Citation of Related Structures:
4R8W, 4UBD - PubMed Abstract:
Effective annual influenza vaccination requires frequent changes in vaccine composition due to both antigenic shift for different subtype hemagglutinins (HAs) and antigenic drift in a particular HA. Here we present a broadly neutralizing human monoclonal antibody with an unusual binding modality. The antibody, designated CT149, was isolated from convalescent patients infected with pandemic H1N1 in 2009. CT149 is found to neutralize all tested group 2 and some group 1 influenza A viruses by inhibiting low pH-induced, HA-mediated membrane fusion. It promotes killing of infected cells by Fc-mediated antibody-dependent cellular cytotoxicity and complement-dependent cytotoxicity. X-ray crystallographic data reveal that CT149 binds primarily to the fusion domain in HA2, and the light chain is also largely involved in binding. The epitope recognized by this antibody comprises amino-acid residues from two adjacent protomers of HA. This binding characteristic of CT149 will provide more information to support the design of more potent influenza vaccines.
- 1] CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, No. 1 West Beichen Road, Beijing 100101, China. [2] Center for Influenza Research and Early-warning, Chinese Academy of Sciences, No. 1 West Beichen Road, Beijing 100101, China.
Organizational Affiliation: