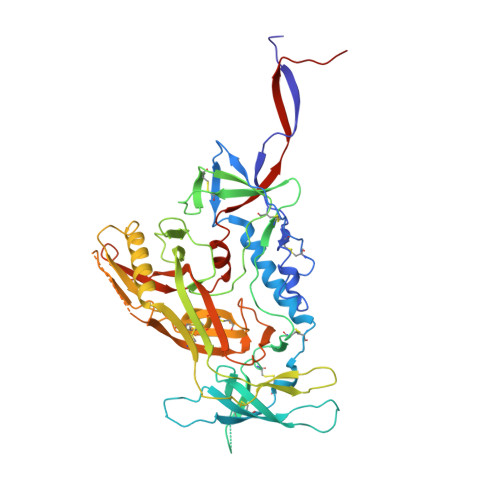
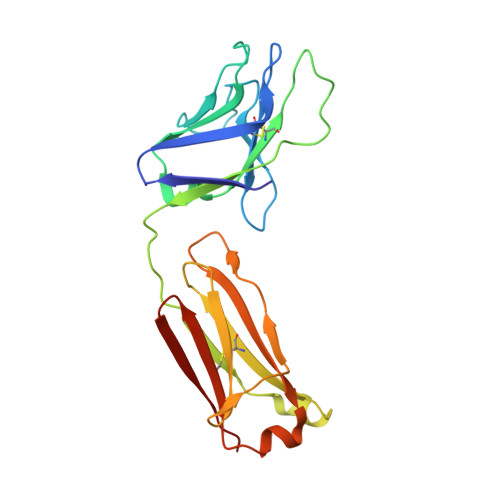
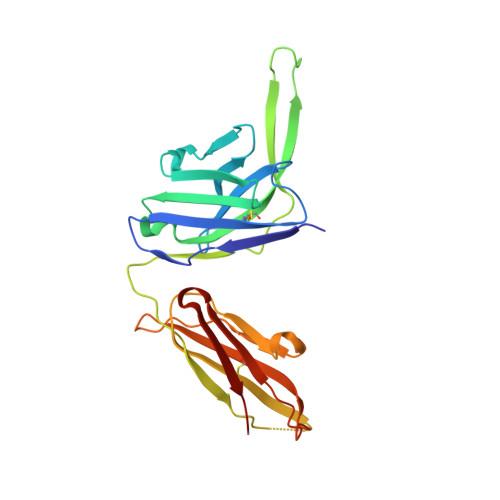
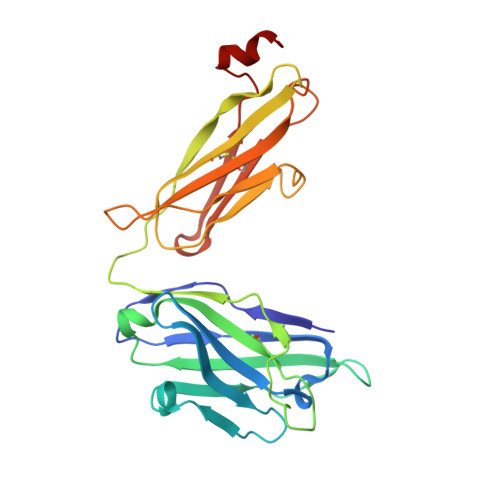
Structure and immune recognition of trimeric pre-fusion HIV-1 Env.
Pancera, M., Zhou, T., Druz, A., Georgiev, I.S., Soto, C., Gorman, J., Huang, J., Acharya, P., Chuang, G.Y., Ofek, G., Stewart-Jones, G.B., Stuckey, J., Bailer, R.T., Joyce, M.G., Louder, M.K., Tumba, N., Yang, Y., Zhang, B., Cohen, M.S., Haynes, B.F., Mascola, J.R., Morris, L., Munro, J.B., Blanchard, S.C., Mothes, W., Connors, M., Kwong, P.D.(2014) Nature 514: 455-461
- PubMed: 25296255
- DOI: https://doi.org/10.1038/nature13808
- Primary Citation of Related Structures:
4TVP - PubMed Abstract:
The human immunodeficiency virus type 1 (HIV-1) envelope (Env) spike, comprising three gp120 and three gp41 subunits, is a conformational machine that facilitates HIV-1 entry by rearranging from a mature unliganded state, through receptor-bound intermediates, to a post-fusion state. As the sole viral antigen on the HIV-1 virion surface, Env is both the target of neutralizing antibodies and a focus of vaccine efforts. Here we report the structure at 3.5 Å resolution for an HIV-1 Env trimer captured in a mature closed state by antibodies PGT122 and 35O22. This structure reveals the pre-fusion conformation of gp41, indicates rearrangements needed for fusion activation, and defines parameters of immune evasion and immune recognition. Pre-fusion gp41 encircles amino- and carboxy-terminal strands of gp120 with four helices that form a membrane-proximal collar, fastened by insertion of a fusion peptide-proximal methionine into a gp41-tryptophan clasp. Spike rearrangements required for entry involve opening the clasp and expelling the termini. N-linked glycosylation and sequence-variable regions cover the pre-fusion closed spike; we used chronic cohorts to map the prevalence and location of effective HIV-1-neutralizing responses, which were distinguished by their recognition of N-linked glycan and tolerance for epitope-sequence variation.
- Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland 20892, USA.
Organizational Affiliation: