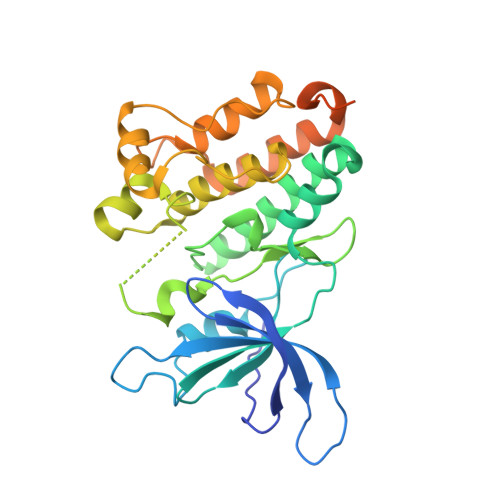
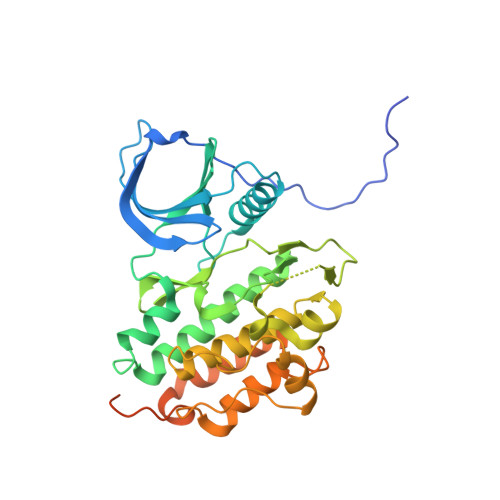
Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations.
Littlefield, P., Liu, L., Mysore, V., Shan, Y., Shaw, D.E., Jura, N.(2014) Sci Signal 7: ra114-ra114
- PubMed: 25468994
- DOI: https://doi.org/10.1126/scisignal.2005786
- Primary Citation of Related Structures:
4RIW, 4RIX, 4RIY - PubMed Abstract:
The human epidermal growth factor receptor (HER) tyrosine kinases homo- and heterodimerize to activate downstream signaling pathways. HER3 is a catalytically impaired member of the HER family that contributes to the development of several human malignancies and is mutated in a subset of cancers. HER3 signaling depends on heterodimerization with a catalytically active partner, in particular epidermal growth factor receptor (EGFR) (the founding family member, also known as HER1) or HER2. The activity of homodimeric complexes of catalytically active HER family members depends on allosteric activation between the two kinase domains. To determine the structural basis for HER3 signaling through heterodimerization with a catalytically active HER family member, we solved the crystal structure of the heterodimeric complex formed by the isolated kinase domains of EGFR and HER3. The structure showed HER3 as an allosteric activator of EGFR and revealed a conserved role of the allosteric mechanism in activation of HER family members through heterodimerization. To understand the effects of cancer-associated HER3 mutations at the molecular level, we solved the structures of two kinase domains of HER3 mutants, each in a heterodimeric complex with the kinase domain of EGFR. These structures, combined with biochemical analysis and molecular dynamics simulations, indicated that the cancer-associated HER3 mutations enhanced the allosteric activator function of HER3 by redesigning local interactions at the dimerization interface.
- Cardiovascular Research Institute, University of California San Francisco, San Francisco, CA 94158, USA.
Organizational Affiliation: