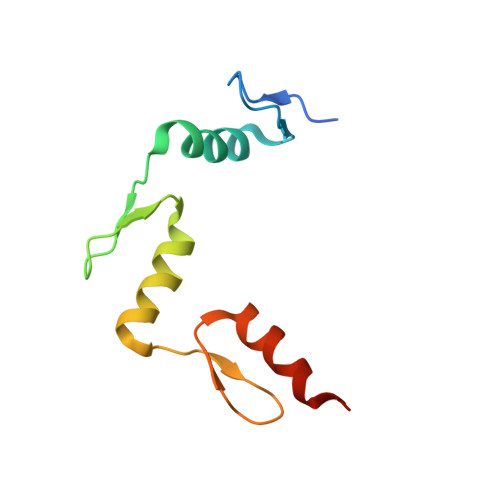
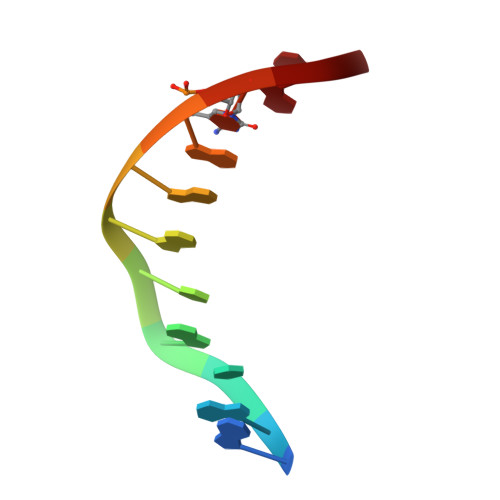
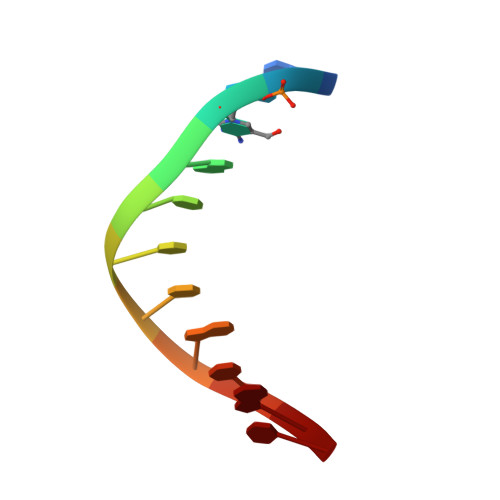
Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Hashimoto, H., Olanrewaju, Y.O., Zheng, Y., Wilson, G.G., Zhang, X., Cheng, X.(2014) Genes Dev 28: 2304-2313
- PubMed: 25258363
- DOI: https://doi.org/10.1101/gad.250746.114
- Primary Citation of Related Structures:
4R2A, 4R2C, 4R2D, 4R2E, 4R2P, 4R2Q, 4R2R, 4R2S - PubMed Abstract:
In mammalian DNA, cytosine occurs in several chemical forms, including unmodified cytosine (C), 5-methylcytosine (5 mC), 5-hydroxymethylcytosine (5 hmC), 5-formylcytosine (5 fC), and 5-carboxylcytosine (5 caC). 5 mC is a major epigenetic signal that acts to regulate gene expression. 5 hmC, 5 fC, and 5 caC are oxidized derivatives that might also act as distinct epigenetic signals. We investigated the response of the zinc finger DNA-binding domains of transcription factors early growth response protein 1 (Egr1) and Wilms tumor protein 1 (WT1) to different forms of modified cytosine within their recognition sequence, 5'-GCG(T/G)GGGCG-3'. Both displayed high affinity for the sequence when C or 5 mC was present and much reduced affinity when 5 hmC or 5 fC was present, indicating that they differentiate primarily oxidized C from unoxidized C, rather than methylated C from unmethylated C. 5 caC affected the two proteins differently, abolishing binding by Egr1 but not by WT1. We ascribe this difference to electrostatic interactions in the binding sites. In Egr1, a negatively charged glutamate conflicts with the negatively charged carboxylate of 5 caC, whereas the corresponding glutamine of WT1 interacts with this group favorably. Our analyses shows that zinc finger proteins (and their splice variants) can respond in modulated ways to alternative modifications within their binding sequence.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, Georgia 30322, USA;
Organizational Affiliation: