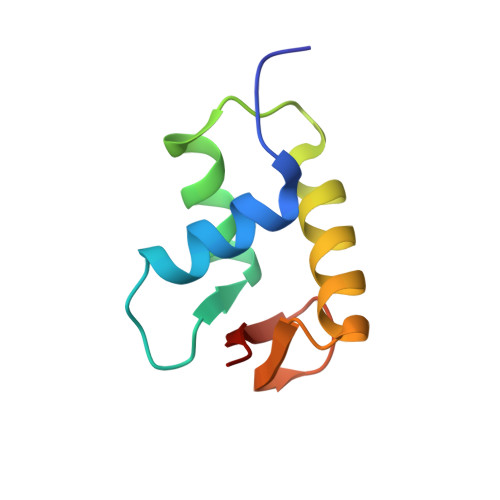
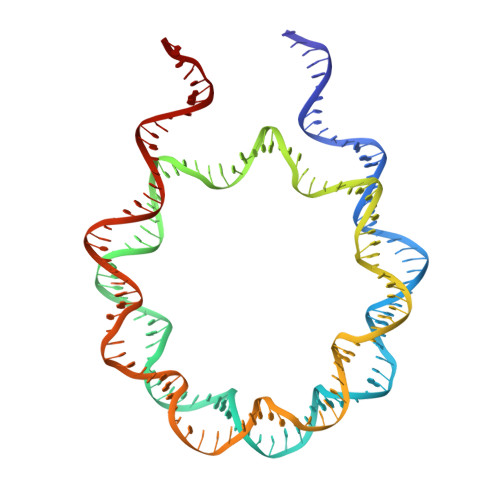
Structural Mechanisms of Nucleosome Recognition by Linker Histones.
Zhou, B.R., Jiang, J., Feng, H., Ghirlando, R., Xiao, T.S., Bai, Y.(2015) Mol Cell 33 Suppl 1: 2-3
- PubMed: 26212454
- DOI: https://doi.org/10.1016/j.molcel.2015.06.025
- Primary Citation of Related Structures:
4QLC - PubMed Abstract:
Linker histones bind to the nucleosome and regulate the structure of chromatin and gene expression. Despite more than three decades of effort, the structural basis of nucleosome recognition by linker histones remains elusive. Here, we report the crystal structure of the globular domain of chicken linker histone H5 in complex with the nucleosome at 3.5 Å resolution, which is validated using nuclear magnetic resonance spectroscopy. The globular domain sits on the dyad of the nucleosome and interacts with both DNA linkers. Our structure integrates results from mutation analyses and previous cross-linking and fluorescence recovery after photobleach experiments, and it helps resolve the long debate on structural mechanisms of nucleosome recognition by linker histones. The on-dyad binding mode of the H5 globular domain is different from the recently reported off-dyad binding mode of Drosophila linker histone H1. We demonstrate that linker histones with different binding modes could fold chromatin to form distinct higher-order structures.
- Laboratory of Biochemistry and Molecular Biology, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: