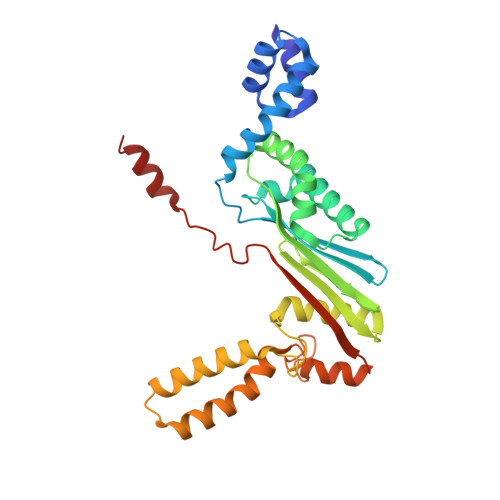
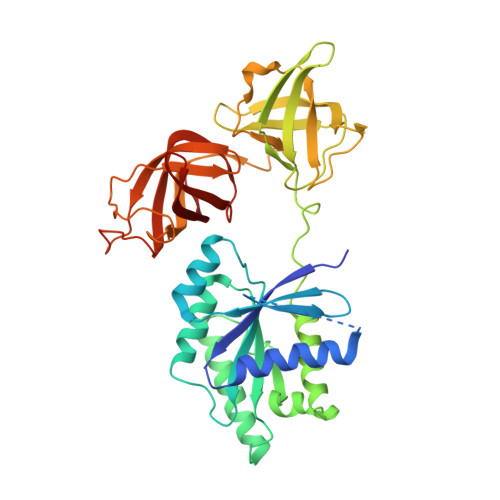
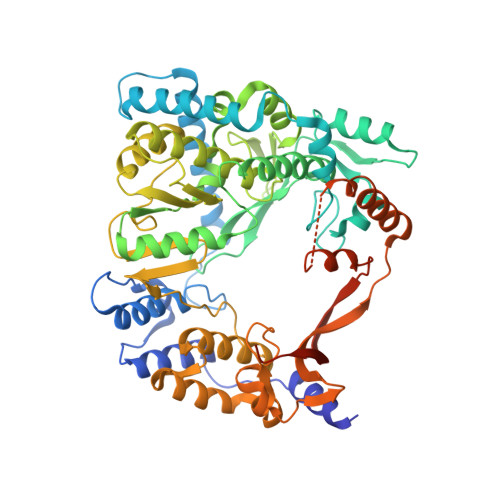
Molecular insights into replication initiation by Q beta replicase using ribosomal protein S1.
Takeshita, D., Yamashita, S., Tomita, K.(2014) Nucleic Acids Res 42: 10809-10822
- PubMed: 25122749
- DOI: https://doi.org/10.1093/nar/gku745
- Primary Citation of Related Structures:
4Q7J - PubMed Abstract:
Ribosomal protein S1, consisting of six contiguous OB-folds, is the largest ribosomal protein and is essential for translation initiation in Escherichia coli. S1 is also one of the three essential host-derived subunits of Qβ replicase, together with EF-Tu and EF-Ts, for Qβ RNA replication in E. coli. We analyzed the crystal structure of Qβ replicase, consisting of the virus-encoded RNA-dependent RNA polymerase (β-subunit), EF-Tu, EF-Ts and the N-terminal half of S1, which is capable of initiating Qβ RNA replication. Structural and biochemical studies revealed that the two N-terminal OB-folds of S1 anchor S1 onto the β-subunit, and the third OB-fold is mobile and protrudes beyond the surface of the β-subunit. The third OB-fold mainly interacts with a specific RNA fragment derived from the internal region of Qβ RNA, and its RNA-binding ability is required for replication initiation of Qβ RNA. Thus, the third mobile OB-fold of S1, which is spatially anchored near the surface of the β-subunit, primarily recruits the Qβ RNA toward the β-subunit, leading to the specific and efficient replication initiation of Qβ RNA, and S1 functions as a replication initiation factor, beyond its established function in protein synthesis.
- Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Tsukuba, Ibaraki 305-8566, Japan.
Organizational Affiliation: