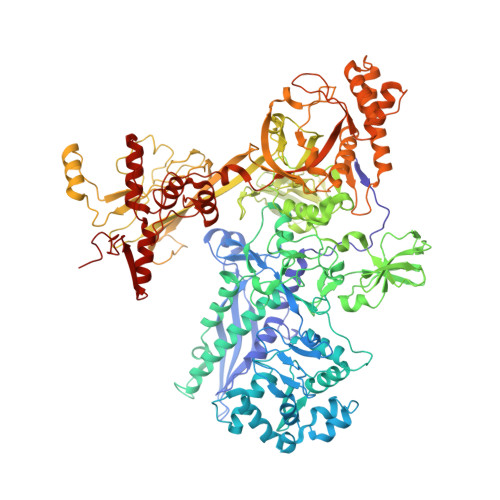
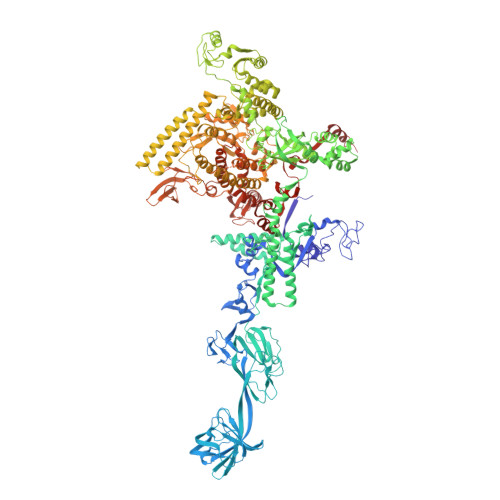
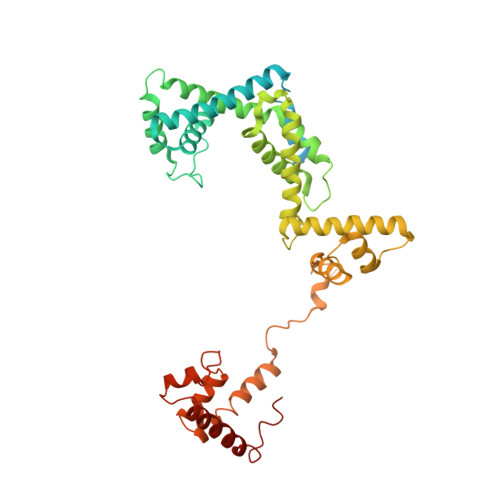
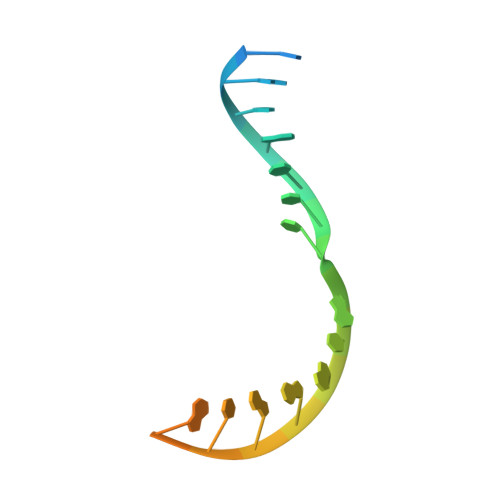
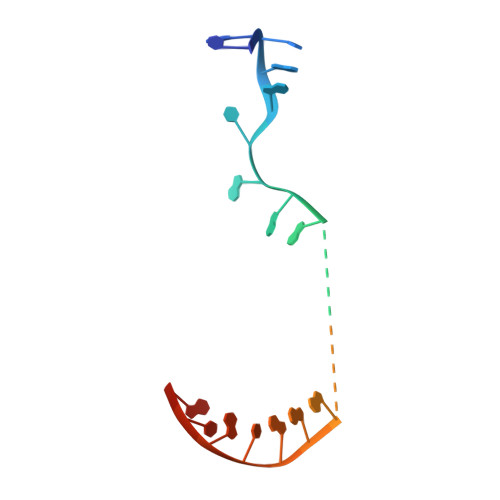
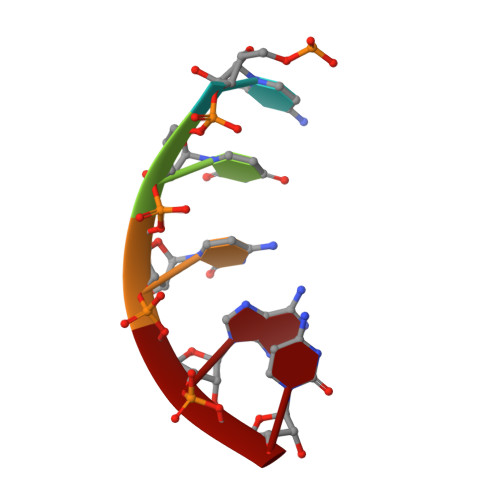
Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme.
Basu, R.S., Warner, B.A., Molodtsov, V., Pupov, D., Esyunina, D., Fernandez-Tornero, C., Kulbachinskiy, A., Murakami, K.S.(2014) J Biological Chem 289: 24549-24559
- PubMed: 24973216
- DOI: https://doi.org/10.1074/jbc.M114.584037
- Primary Citation of Related Structures:
4Q4Z, 4Q5S - PubMed Abstract:
The bacterial RNA polymerase (RNAP) holoenzyme containing σ factor initiates transcription at specific promoter sites by de novo RNA priming, the first step of RNA synthesis where RNAP accepts two initiating ribonucleoside triphosphates (iNTPs) and performs the first phosphodiester bond formation. We present the structure of de novo transcription initiation complex that reveals unique contacts of the iNTPs bound at the transcription start site with the template DNA and also with RNAP and demonstrate the importance of these contacts for transcription initiation. To get further insight into the mechanism of RNA priming, we determined the structure of initially transcribing complex of RNAP holoenzyme with 6-mer RNA, obtained by in crystallo transcription approach. The structure highlights RNAP-RNA contacts that stabilize the short RNA transcript in the active site and demonstrates that the RNA 5'-end displaces σ region 3.2 from its position near the active site, which likely plays a key role in σ ejection during the initiation-to-elongation transition. Given the structural conservation of the RNAP active site, the mechanism of de novo RNA priming appears to be conserved in all cellular RNAPs.
- From the Department of Biochemistry and Molecular Biology, The Center for RNA Molecular Biology, The Pennsylvania State University, University Park, Pennsylvania 16802.
Organizational Affiliation: