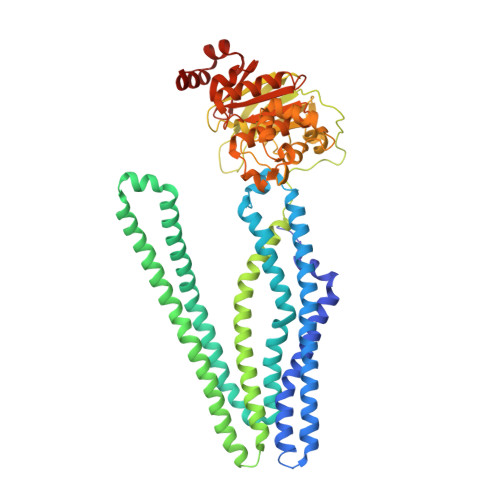
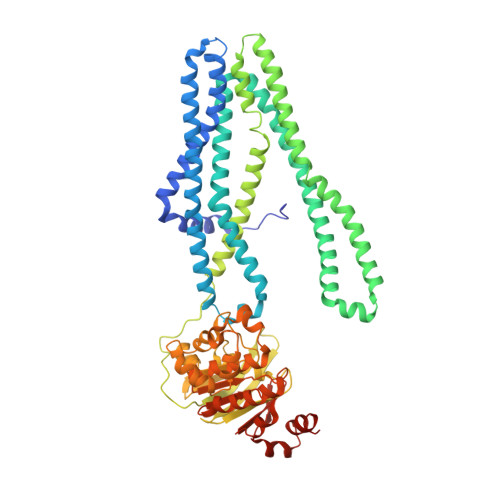
Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter.
Hohl, M., Hurlimann, L.M., Bohm, S., Schoppe, J., Grutter, M.G., Bordignon, E., Seeger, M.A.(2014) Proc Natl Acad Sci U S A 111: 11025-11030
- PubMed: 25030449
- DOI: https://doi.org/10.1073/pnas.1400485111
- Primary Citation of Related Structures:
4Q4A, 4Q4H, 4Q4J - PubMed Abstract:
ATP binding cassette (ABC) transporters mediate vital transport processes in every living cell. ATP hydrolysis, which fuels transport, displays positive cooperativity in numerous ABC transporters. In particular, heterodimeric ABC exporters exhibit pronounced allosteric coupling between a catalytically impaired degenerate site, where nucleotides bind tightly, and a consensus site, at which ATP is hydrolyzed in every transport cycle. Whereas the functional phenomenon of cooperativity is well described, its structural basis remains poorly understood. Here, we present the apo structure of the heterodimeric ABC exporter TM287/288 and compare it to the previously solved structure with adenosine 5'-(β,γ-imido)triphosphate (AMP-PNP) bound at the degenerate site. In contrast to other ABC exporter structures, the nucleotide binding domains (NBDs) of TM287/288 remain in molecular contact even in the absence of nucleotides, and the arrangement of the transmembrane domains (TMDs) is not influenced by AMP-PNP binding, a notion confirmed by double electron-electron resonance (DEER) measurements. Nucleotide binding at the degenerate site results in structural rearrangements, which are transmitted to the consensus site via two D-loops located at the NBD interface. These loops owe their name from a highly conserved aspartate and are directly connected to the catalytically important Walker B motif. The D-loop at the degenerate site ties the NBDs together even in the absence of nucleotides and substitution of its aspartate by alanine is well-tolerated. By contrast, the D-loop of the consensus site is flexible and the aspartate to alanine mutation and conformational restriction by cross-linking strongly reduces ATP hydrolysis and substrate transport.
- Department of Biochemistry, University of Zurich, CH-8057 Zurich, Switzerland;Institute of Medical Microbiology, University of Zurich, CH-8006 Zurich, Switzerland;
Organizational Affiliation: