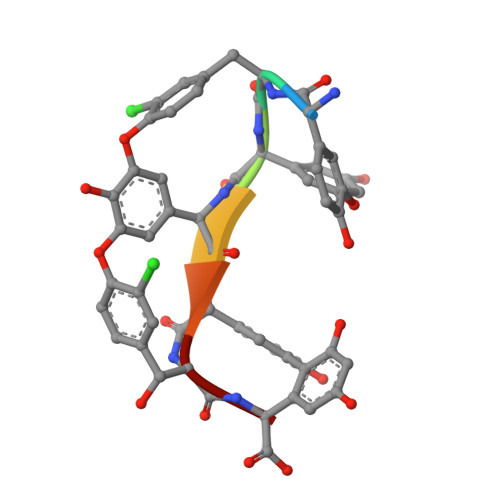
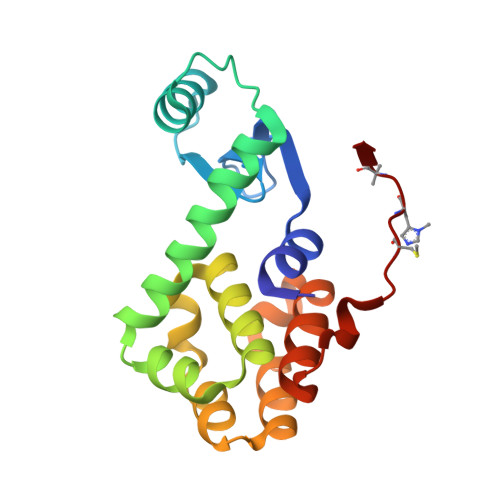
X-ray Crystal Structure of Teicoplanin A2-2 Bound to a Catalytic Peptide Sequence via the Carrier Protein Strategy.
Han, S., Le, B.V., Hajare, H.S., Baxter, R.H., Miller, S.J.(2014) J Org Chem 79: 8550-8556
- PubMed: 25147913
- DOI: https://doi.org/10.1021/jo501625f
- Primary Citation of Related Structures:
4PJZ, 4PK0 - PubMed Abstract:
We report the X-ray crystal structure of a site-selective peptide catalyst moiety and teicoplanin A2-2 complex. The expressed protein ligation technique was used to couple T4 lysozyme (T4L) and a synthetic peptide catalyst responsible for the selective phosphorylation of the N-acetylglucosamine sugar in a teicoplanin A2-2 derivative. The T4L-Pmh-dPro-Aib-dAla-dAla construct was crystallized in the presence of teicoplanin A2-2. The resulting 2.3 Å resolution protein-peptide-teicoplanin complex crystal structure revealed that the nucleophilic nitrogen of N-methylimidazole in the Pmh residue is in closer proximity (7.6 Å) to the N-acetylglucosamine than the two other sugar rings present in teicoplanin (9.3 and 20.3 Å, respectively). This molecular arrangement is consistent with the observed selectivity afforded by the peptide-based catalyst when it is applied to a site-selective phosphorylation reaction involving a teicoplanin A2-2 derivative.
- Department of Chemistry, Yale University , New Haven, Connecticut 06511, United States.
Organizational Affiliation: