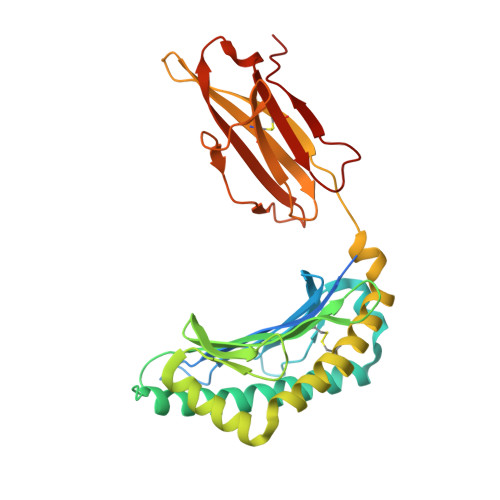
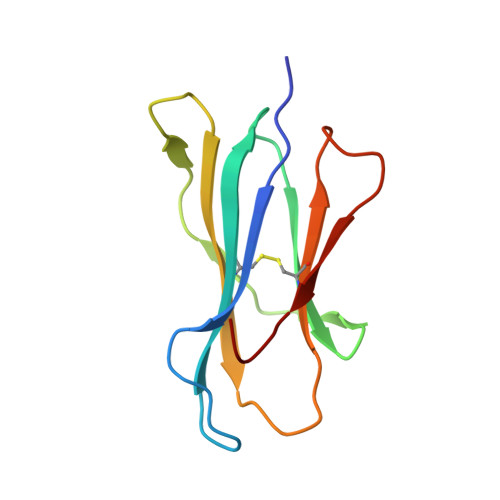
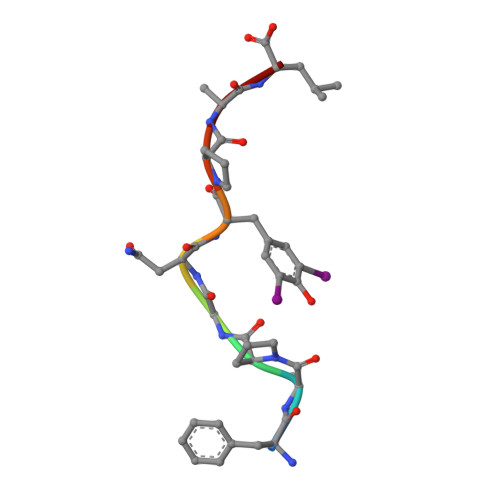
The first step of peptide selection in antigen presentation by MHC class I molecules.
Garstka, M.A., Fish, A., Celie, P.H., Joosten, R.P., Janssen, G.M., Berlin, I., Hoppes, R., Stadnik, M., Janssen, L., Ovaa, H., van Veelen, P.A., Perrakis, A., Neefjes, J.(2015) Proc Natl Acad Sci U S A 112: 1505-1510
- PubMed: 25605945
- DOI: https://doi.org/10.1073/pnas.1416543112
- Primary Citation of Related Structures:
4PG9, 4PGB, 4PGC, 4PGD, 4PGE - PubMed Abstract:
MHC class I molecules present a variable but limited repertoire of antigenic peptides for T-cell recognition. Understanding how peptide selection is achieved requires mechanistic insights into the interactions between the MHC I and candidate peptides. We find that, at first encounter, MHC I H-2K(b) considers a wide range of peptides, including those with expanded N termini and unfitting anchor residues. Discrimination occurs in the second step, when noncanonical peptides dissociate with faster exchange rates. This second step exhibits remarkable temperature sensitivity, as illustrated by numerous noncanonical peptides presented by H-2K(b) in cells cultured at 26 °C relative to 37 °C. Crystallographic analyses of H-2K(b)-peptide complexes suggest that a conformational adaptation of H-2K(b) drives the decisive step in peptide selection. We propose that MHC class I molecules consider initially a large peptide pool, subsequently refined by a temperature-sensitive induced-fit mechanism to retain the canonical peptide repertoire.
- Departments of Cell Biology and.
Organizational Affiliation: