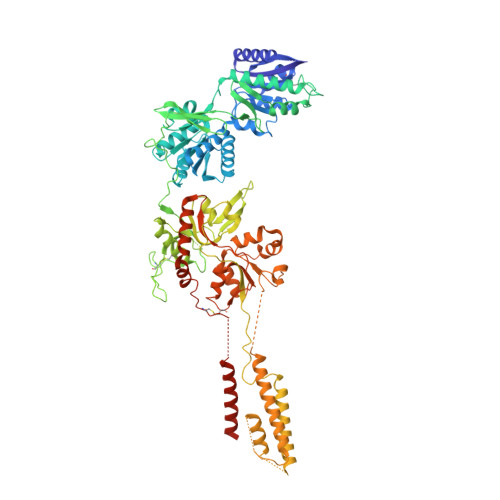
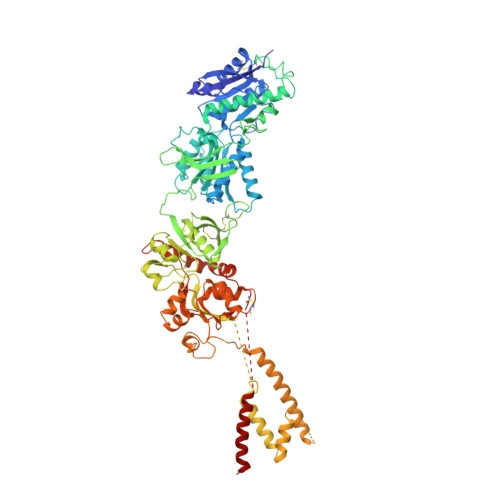
Crystal structure of a heterotetrameric NMDA receptor ion channel.
Karakas, E., Furukawa, H.(2014) Science 344: 992-997
- PubMed: 24876489
- DOI: https://doi.org/10.1126/science.1251915
- Primary Citation of Related Structures:
4PE5 - PubMed Abstract:
N-Methyl-D-aspartate (NMDA) receptors belong to the family of ionotropic glutamate receptors, which mediate most excitatory synaptic transmission in mammalian brains. Calcium permeation triggered by activation of NMDA receptors is the pivotal event for initiation of neuronal plasticity. Here, we show the crystal structure of the intact heterotetrameric GluN1-GluN2B NMDA receptor ion channel at 4 angstroms. The NMDA receptors are arranged as a dimer of GluN1-GluN2B heterodimers with the twofold symmetry axis running through the entire molecule composed of an amino terminal domain (ATD), a ligand-binding domain (LBD), and a transmembrane domain (TMD). The ATD and LBD are much more highly packed in the NMDA receptors than non-NMDA receptors, which may explain why ATD regulates ion channel activity in NMDA receptors but not in non-NMDA receptors.
- Cold Spring Harbor Laboratory, W. M. Keck Structural Biology Laboratory, One Bungtown Road, Cold Spring Harbor, NY 11724, USA.
Organizational Affiliation: