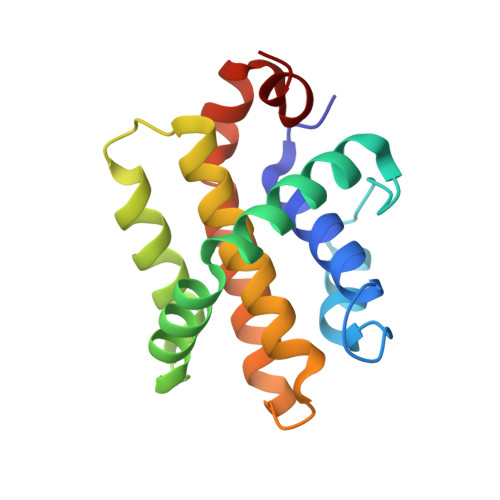
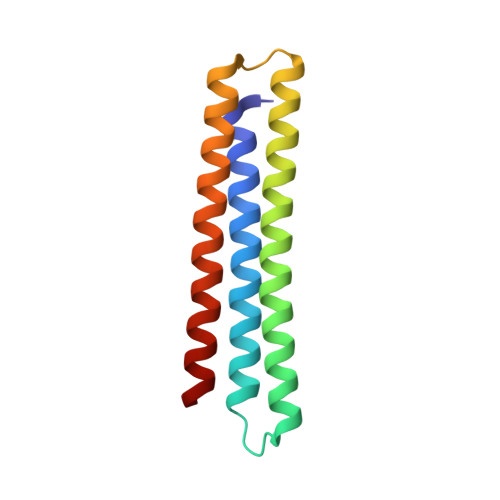
A computationally designed inhibitor of an epstein-barr viral bcl-2 protein induces apoptosis in infected cells.
Procko, E., Berguig, G.Y., Shen, B.W., Song, Y., Frayo, S., Convertine, A.J., Margineantu, D., Booth, G., Correia, B.E., Cheng, Y., Schief, W.R., Hockenbery, D.M., Press, O.W., Stoddard, B.L., Stayton, P.S., Baker, D.(2014) Cell 157: 1644-1656
- PubMed: 24949974
- DOI: https://doi.org/10.1016/j.cell.2014.04.034
- Primary Citation of Related Structures:
4OYD - PubMed Abstract:
Because apoptosis of infected cells can limit virus production and spread, some viruses have co-opted prosurvival genes from the host. This includes the Epstein-Barr virus (EBV) gene BHRF1, a homolog of human Bcl-2 proteins that block apoptosis and are associated with cancer. Computational design and experimental optimization were used to generate a novel protein called BINDI that binds BHRF1 with picomolar affinity. BINDI recognizes the hydrophobic cleft of BHRF1 in a manner similar to other Bcl-2 protein interactions but makes many additional contacts to achieve exceptional affinity and specificity. BINDI induces apoptosis in EBV-infected cancer lines, and when delivered with an antibody-targeted intracellular delivery carrier, BINDI suppressed tumor growth and extended survival in a xenograft disease model of EBV-positive human lymphoma. High-specificity-designed proteins that selectively kill target cells may provide an advantage over the toxic compounds used in current generation antibody-drug conjugates.
- Department of Biochemistry, University of Washington, Seattle, WA 98195, USA.
Organizational Affiliation: