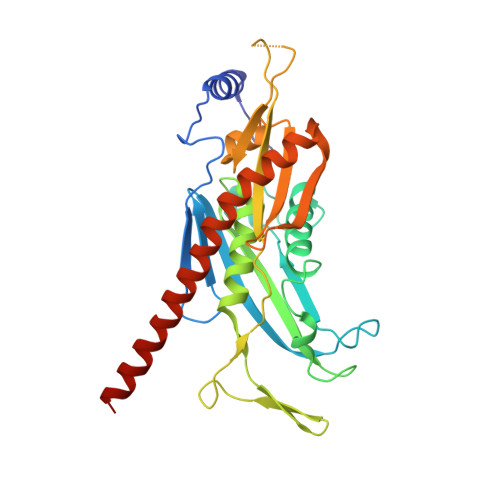
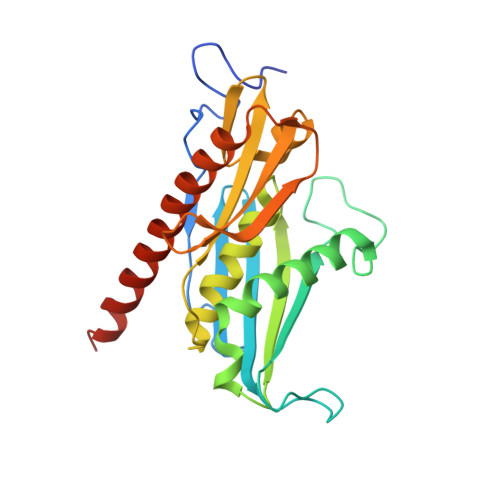
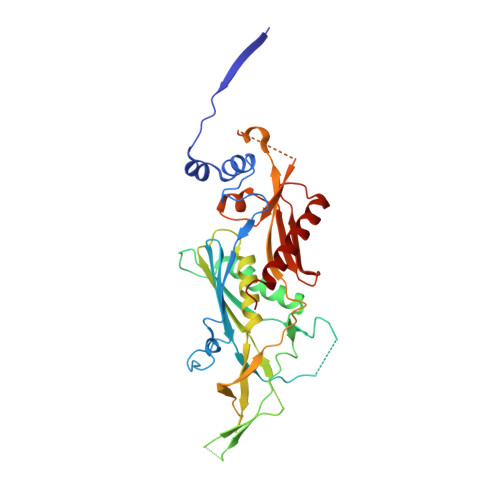
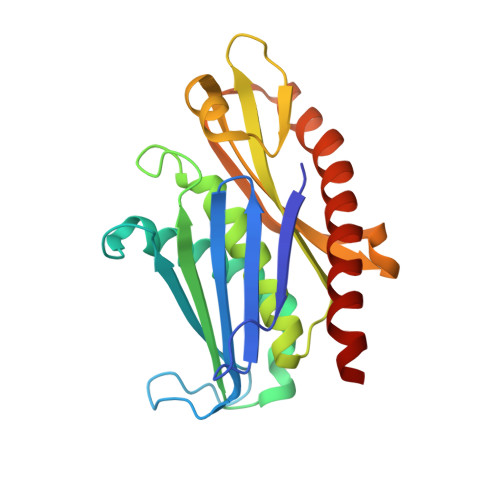
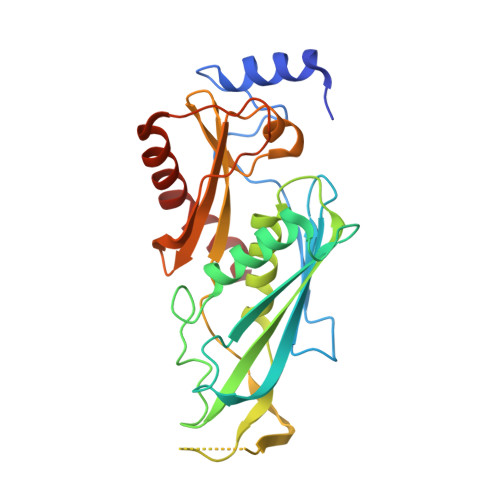
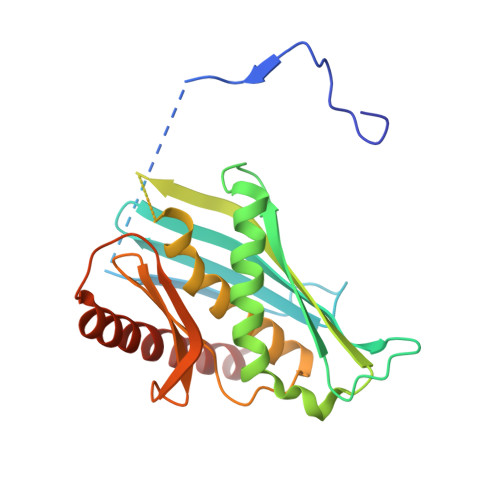
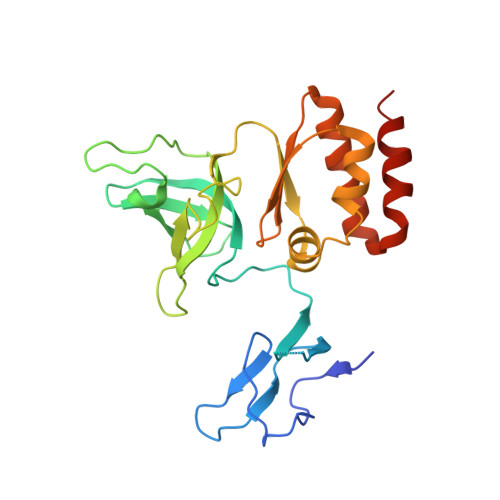
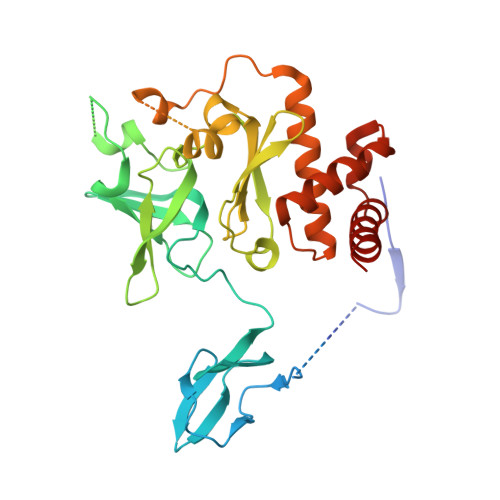
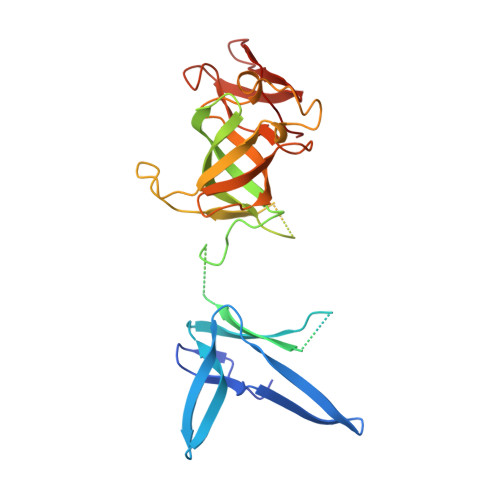
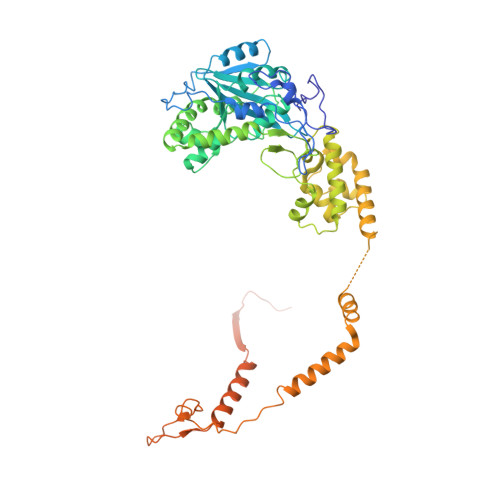
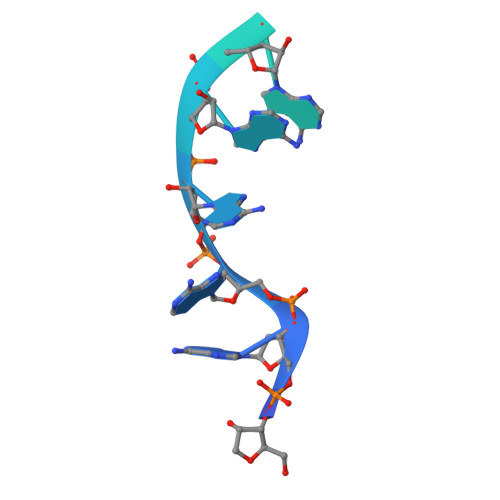
Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA.
Wasmuth, E.V., Januszyk, K., Lima, C.D.(2014) Nature 511: 435-439
- PubMed: 25043052
- DOI: https://doi.org/10.1038/nature13406
- Primary Citation of Related Structures:
4OO1 - PubMed Abstract:
The eukaryotic RNA exosome processes and degrades RNA by directing substrates to the distributive or processive 3' to 5' exoribonuclease activities of Rrp6 or Rrp44, respectively. The non-catalytic nine-subunit exosome core (Exo9) features a prominent central channel. Although RNA can pass through the channel to engage Rrp44, it is not clear how RNA is directed to Rrp6 or whether Rrp6 uses the central channel. Here we report a 3.3 Å crystal structure of a ten-subunit RNA exosome complex from Saccharomyces cerevisiae composed of the Exo9 core and Rrp6 bound to single-stranded poly(A) RNA. The Rrp6 catalytic domain rests on top of the Exo9 S1/KH ring above the central channel, the RNA 3' end is anchored in the Rrp6 active site, and the remaining RNA traverses the S1/KH ring in an opposite orientation to that observed in a structure of a Rrp44-containing exosome complex. Solution studies with human and yeast RNA exosome complexes suggest that the RNA path to Rrp6 is conserved and dependent on the integrity of the S1/KH ring. Although path selection to Rrp6 or Rrp44 is stochastic in vitro, the fate of a particular RNA may be determined in vivo by the manner in which cofactors present RNA to the RNA exosome.
- 1] Structural Biology Program, Sloan-Kettering Institute, 1275 York Avenue, New York, New York 10065, USA [2] Louis V. Gerstner Jr. Graduate School of Biomedical Sciences, Sloan-Kettering Institute, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, New York 10065, USA.
Organizational Affiliation: