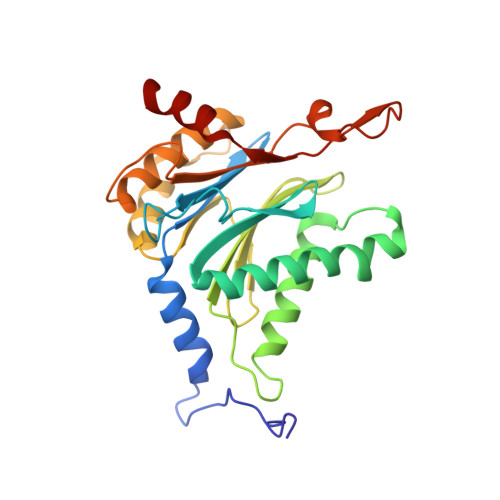
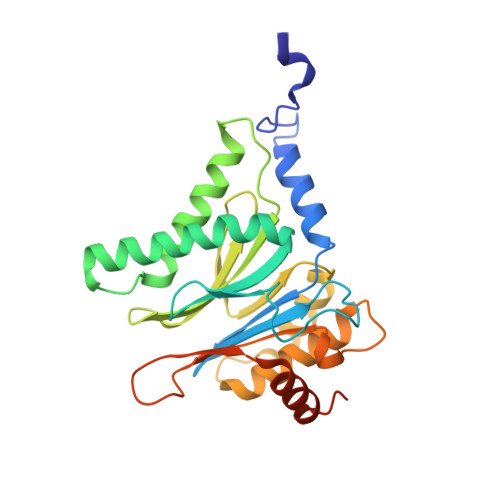
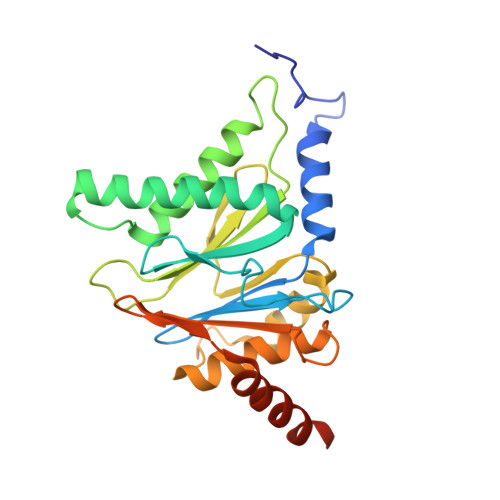
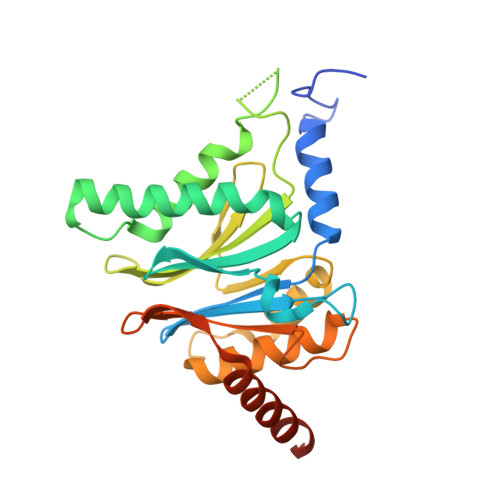
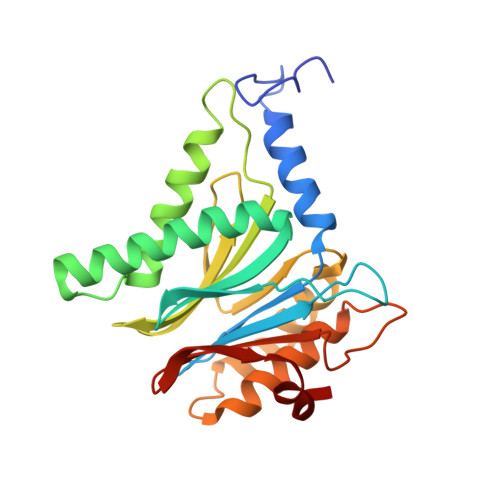
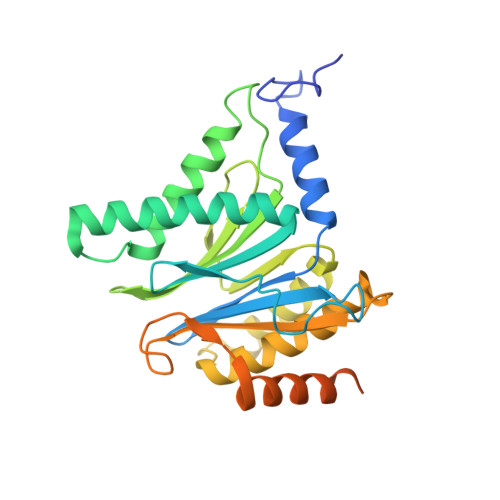
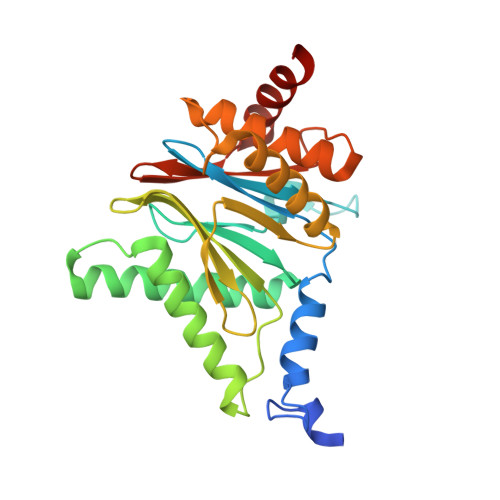
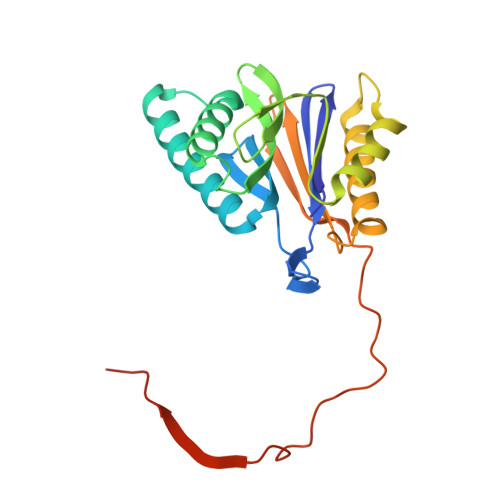
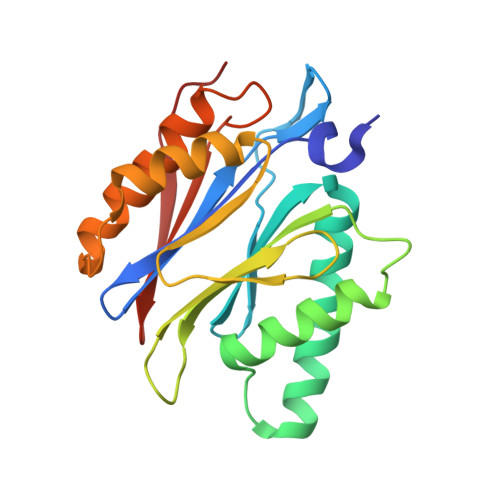
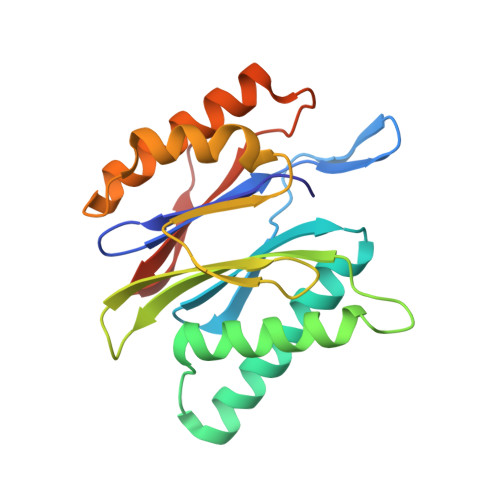
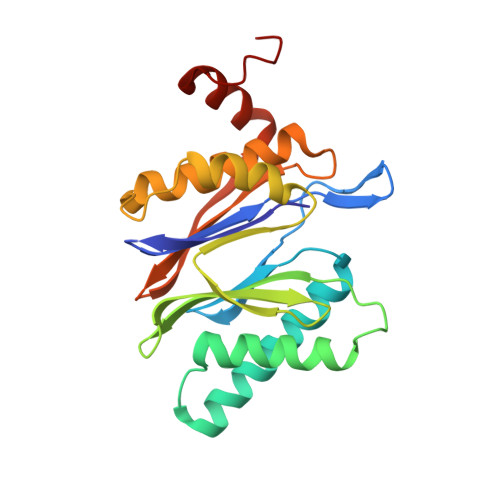
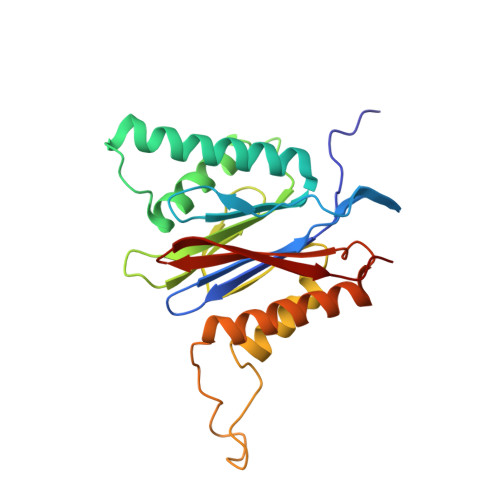
Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Stein, M.L., Cui, H., Beck, P., Dubiella, C., Voss, C., Kruger, A., Schmidt, B., Groll, M.(2014) Angew Chem Int Ed Engl 53: 1679-1683
- PubMed: 24403024
- DOI: https://doi.org/10.1002/anie.201308984
- Primary Citation of Related Structures:
4NNN, 4NNW, 4NO1, 4NO6, 4NO8, 4NO9 - PubMed Abstract:
The ubiquitin-proteasome system (UPS) has been successfully targeted by both academia and the pharmaceutical industry for oncological and immunological applications. Typical proteasome inhibitors are based on a peptidic backbone endowed with an electrophilic C-terminus by which they react with the active proteolytic sites. Although the peptide moiety has attracted much attention in terms of subunit selectivity, the target specificity and biological stability of the compounds are largely determined by the reactive warheads. In this study, we have carried out a systematic investigation of described electrophiles by a combination of in vitro, in vivo, and structural methods in order to disclose the implications of altered functionality and chemical reactivity. Thereby, we were able to introduce and characterize the class of α-ketoamides as the most potent reversible inhibitors with possible applications for the therapy of solid tumors as well as autoimmune disorders.
- Center for Integrated Protein Science at the Department Chemie, Lehrstuhl für Biochemie, TU München (Germany).
Organizational Affiliation: