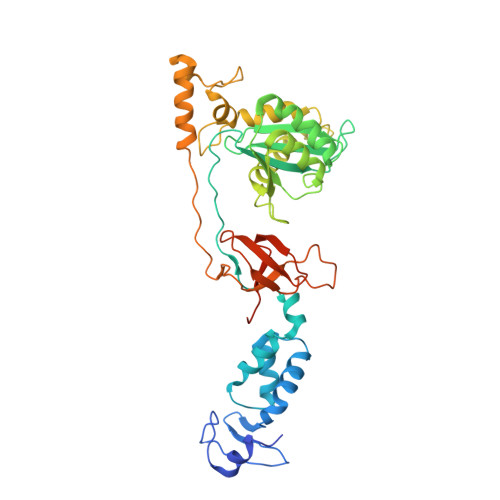
2-Hydroxyisoquinoline-1,3(2H,4H)-diones (HIDs), novel inhibitors of HIV integrase with a high barrier to resistance.
Desimmie, B.A., Demeulemeester, J., Suchaud, V., Taltynov, O., Billamboz, M., Lion, C., Bailly, F., Strelkov, S.V., Debyser, Z., Cotelle, P., Christ, F.(2013) ACS Chem Biol 8: 1187-1194
- PubMed: 23517458
- DOI: https://doi.org/10.1021/cb4000426
- Primary Citation of Related Structures:
4IKF - PubMed Abstract:
Clinical HIV-1 integrase (IN) strand transfer inhibitors (INSTIs) potently inhibit viral replication with a dramatic drop in viral load. However, the emergence of resistance to these drugs underscores the need to develop next-generation IN catalytic site inhibitors with improved resistance profiles. Here, we present a novel candidate IN inhibitor, MB-76, a 2-hydroxyisoquinoline-1,3(2H,4H)-dione (HID) derivative. MB-76 potently blocks HIV integration and is active against a panel of wild-type as well as raltegravir-resistant HIV-1 variants. The lack of cross-resistance with other INSTIs and the absence of resistance selection in cell culture indicate the potential of HID derivatives compared to previous INSTIs. A crystal structure of MB-76 bound to the wild-type prototype foamy virus intasome reveals an overall binding mode similar to that of INSTIs. Its compact scaffold displays all three Mg(2+) chelating oxygen atoms from a single ring, ensuring that the only direct contacts with IN are the invariant P214 and Q215 residues of PFV IN (P145 and Q146 for HIV-1 IN, respectively), which may partially explain the difficulty of selecting replicating resistant variants. Moreover, the extended, dolutegravir-like linker connecting the MB-76 metal chelating core and p-fluorobenzyl group can provide additional flexibility in the perturbed active sites of raltegravir-resistant INs. The compound identified represents a potential candidate for further (pre)clinical development as next-generation HIV IN catalytic site inhibitor.
- Molecular Virology and Gene Therapy and §Laboratory for Biocrystallography, KU Leuven , Department of Pharmaceutical and Pharmacological Sciences, Leuven, Belgium.
Organizational Affiliation: