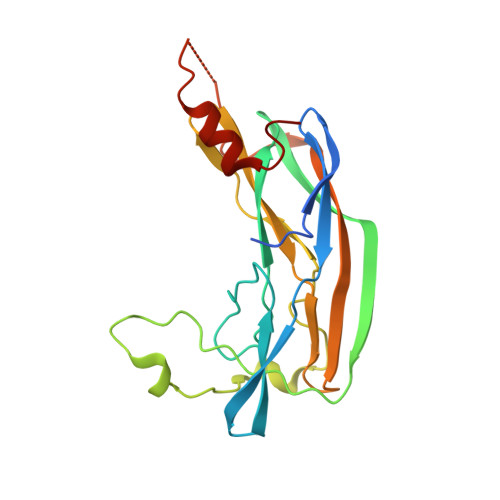
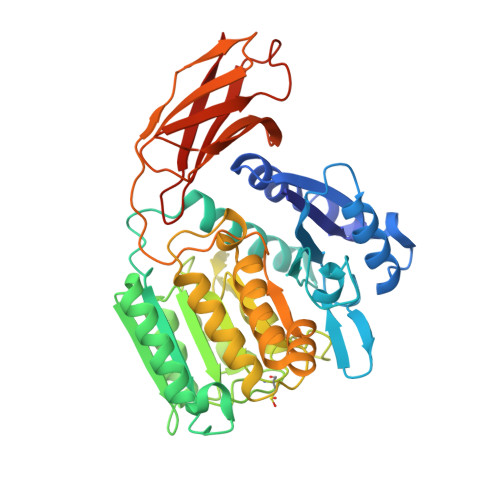
Porphyromonas gingivalis Virulence Factor Gingipain RgpB Shows a Unique Zymogenic Mechanism for Cysteine Peptidases.
de Diego, I., Veillard, F.T., Guevara, T., Potempa, B., Sztukowska, M., Potempa, J., Gomis-Ruth, F.X.(2013) J Biological Chem 288: 14287-14296
- PubMed: 23558682
- DOI: https://doi.org/10.1074/jbc.M112.444927
- Primary Citation of Related Structures:
4IEF - PubMed Abstract:
Zymogenicity is a regulatory mechanism that prevents inadequate catalytic activity in the wrong context. It plays a central role in maintaining microbial virulence factors in an inactive form inside the pathogen until secretion. Among these virulence factors is the cysteine peptidase gingipain B (RgpB), which is the major virulence factor secreted by the periodontopathogen Porphyromonas gingivalis that attacks host vasculature and defense proteins. The structure of the complex between soluble mature RgpB, consisting of a catalytic domain and an immunoglobulin superfamily domain, and its 205-residue N-terminal prodomain, the largest structurally characterized to date for a cysteine peptidase, reveals a novel fold for the prodomain that is distantly related to sugar-binding lectins. It attaches laterally to the catalytic domain through a large concave surface. The main determinant for latency is a surface "inhibitory loop," which approaches the active-site cleft of the enzyme on its non-primed side in a substrate-like manner. It inserts an arginine (Arg(126)) into the S1 pocket, thus matching the substrate specificity of the enzyme. Downstream of Arg(126), the polypeptide leaves the cleft, thereby preventing cleavage. Moreover, the carbonyl group of Arg(126) establishes a very strong hydrogen bond with the co-catalytic histidine, His(440), pulling it away from the catalytic cysteine, Cys(473), and toward Glu(381), which probably plays a role in orienting the side chain of His(440) during catalysis. The present results provide the structural determinants of zymogenic inhibition of RgpB by way of a novel inhibitory mechanism for peptidases in general and open the field for the design of novel inhibitory strategies in the treatment of human periodontal disease.
- Proteolysis Laboratory, Molecular Biology Institute of Barcelona, Spanish Research Council (CSIC), Barcelona Science Park, c/Baldiri Reixac, 15-21, 08028 Barcelona, Catalonia, Spain.
Organizational Affiliation: