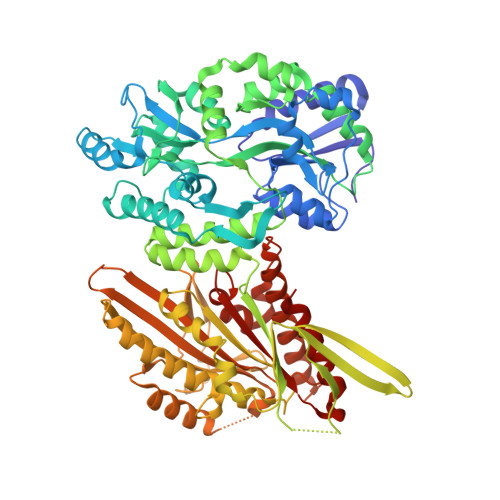
Crystal structure of the Candida albicans Kar3 kinesin motor domain fused to maltose-binding protein.
Delorme, C., Joshi, M., Allingham, J.S.(2012) Biochem Biophys Res Commun 428: 427-432
- PubMed: 23137538
- DOI: https://doi.org/10.1016/j.bbrc.2012.10.101
- Primary Citation of Related Structures:
4H1G - PubMed Abstract:
In the human fungal pathogen Candida albicans, the Kinesin-14 motor protein Kar3 (CaKar3) is critical for normal mitotic division, nuclear fusion during mating, and morphogenic transition from the commensal yeast form to the virulent hyphal form. As a first step towards detailed characterization of this motor of potential medical significance, we have crystallized and determined the X-ray structure of the motor domain of CaKar3 as a maltose-binding protein (MBP) fusion. The structure shows strong conservation of overall motor domain topology to other Kar3 kinesins, but with some prominent differences in one of the motifs that compose the nucleotide-binding pocket and the surface charge distribution. The MBP and Kar3 modules are arranged such that MBP interacts with the Kar3 motor domain core at the same site where the neck linker of conventional kinesins docks during the "ATP state" of the mechanochemical cycle. This site differs from the Kar3 neck-core interface in the recent structure of the ScKar3Vik1 heterodimer. The position of MBP is also completely distinct from the Vik1 subunit in this complex. This may suggest that the site of MBP interaction on the CaKar3 motor domain provides an interface for the neck, or perhaps a partner subunit, at an intermediate state of its motile cycle that has not yet been observed for Kinesin-14 motors.
- Department of Biomedical and Molecular Sciences, Queen's University, Kingston, ON, Canada K7L 3N6.
Organizational Affiliation: