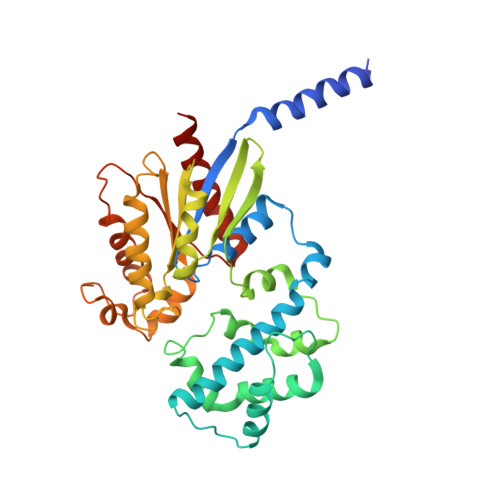
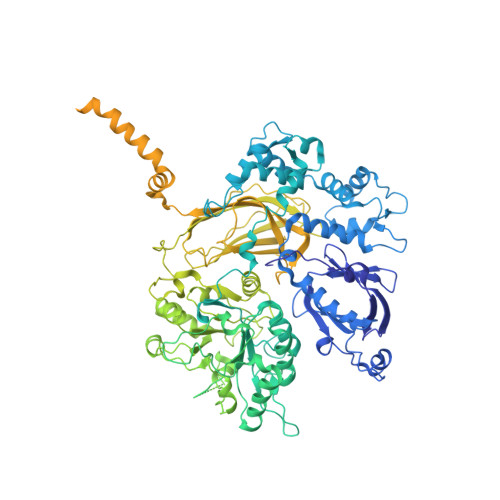
Full-length G alpha (q)-phospholipase C-beta 3 structure reveals interfaces of the C-terminal coiled-coil domain.
Lyon, A.M., Dutta, S., Boguth, C.A., Skiniotis, G., Tesmer, J.J.(2013) Nat Struct Mol Biol 20: 355-362
- PubMed: 23377541
- DOI: https://doi.org/10.1038/nsmb.2497
- Primary Citation of Related Structures:
4GNK - PubMed Abstract:
Phospholipase C-β (PLCβ) is directly activated by Gαq, but the molecular basis for how its distal C-terminal domain (CTD) contributes to maximal activity is poorly understood. Herein we present both the crystal structure and cryo-EM three-dimensional reconstructions of human full-length PLCβ3 in complex with mouse Gαq. The distal CTD forms an extended monomeric helical bundle consisting of three antiparallel segments with structural similarity to membrane-binding bin-amphiphysin-Rvs (BAR) domains. Sequence conservation of the distal CTD suggests putative membrane and protein interaction sites, the latter of which bind the N-terminal helix of Gαq in both the crystal structure and cryo-EM reconstructions. Functional analysis suggests that the distal CTD has roles in membrane targeting and in optimizing the orientation of the catalytic core at the membrane for maximal rates of lipid hydrolysis.
- Life Sciences Institute, University of Michigan, Ann Arbor, Michigan, USA.
Organizational Affiliation: