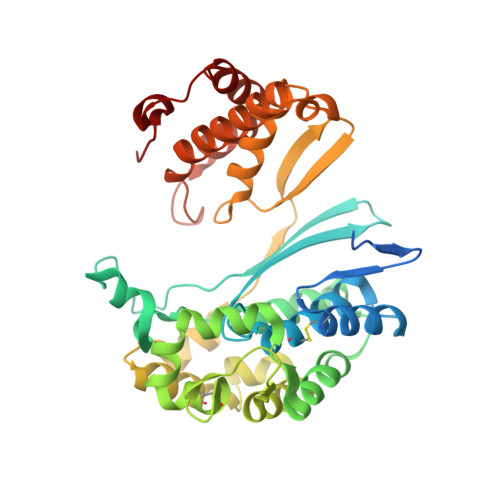
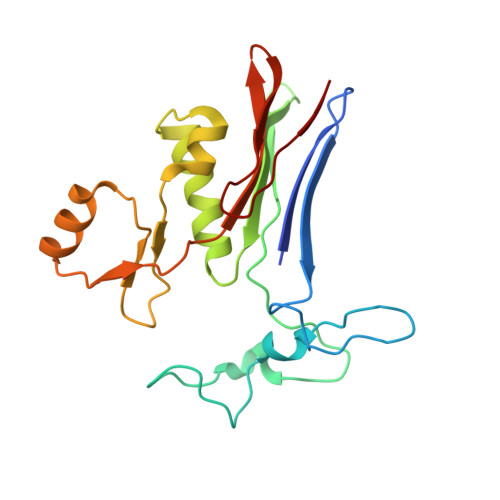
Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
West, M.B., Chen, Y., Wickham, S., Heroux, A., Cahill, K., Hanigan, M.H., Mooers, B.H.(2013) J Biological Chem 288: 31902-31913
- PubMed: 24047895
- DOI: https://doi.org/10.1074/jbc.M113.498139
- Primary Citation of Related Structures:
4GDX, 4GG2 - PubMed Abstract:
The enzyme γ-glutamyltranspeptidase 1 (GGT1) is a conserved member of the N-terminal nucleophile hydrolase family that cleaves the γ-glutamyl bond of glutathione and other γ-glutamyl compounds. In animals, GGT1 is expressed on the surface of the cell and has critical roles in maintaining cysteine levels in the body and regulating intracellular redox status. Expression of GGT1 has been implicated as a potentiator of asthma, cardiovascular disease, and cancer. The rational design of effective inhibitors of human GGT1 (hGGT1) has been delayed by the lack of a reliable structural model. The available crystal structures of several bacterial GGTs have been of limited use due to differences in the catalytic behavior of bacterial and mammalian GGTs. We report the high resolution (1.67 Å) crystal structure of glutamate-bound hGGT1, the first of any eukaryotic GGT. Comparisons of the active site architecture of hGGT1 with those of its bacterial orthologs highlight key differences in the residues responsible for substrate binding, including a bimodal switch in the orientation of the catalytic nucleophile (Thr-381) that is unique to the human enzyme. Compared with several bacterial counterparts, the lid loop in the crystal structure of hGGT1 adopts an open conformation that allows greater access to the active site. The hGGT1 structure also revealed tightly bound chlorides near the catalytic residue that may contribute to catalytic activity. These are absent in the bacterial GGTs. These differences between bacterial and mammalian GGTs and the new structural data will accelerate the development of new therapies for GGT1-dependent diseases.
- From the Departments of Cell Biology and.
Organizational Affiliation: