Unique thrombin inhibition mechanism by anophelin, an anticoagulant from the malaria vector.
Figueiredo, A.C., de Sanctis, D., Gutierrez-Gallego, R., Cereija, T.B., Macedo-Ribeiro, S., Fuentes-Prior, P., Pereira, P.J.(2012) Proc Natl Acad Sci U S A 109: E3649-E3658
- PubMed: 23223529
- DOI: https://doi.org/10.1073/pnas.1211614109
- Primary Citation of Related Structures:
4E05, 4E06 - PubMed Abstract:
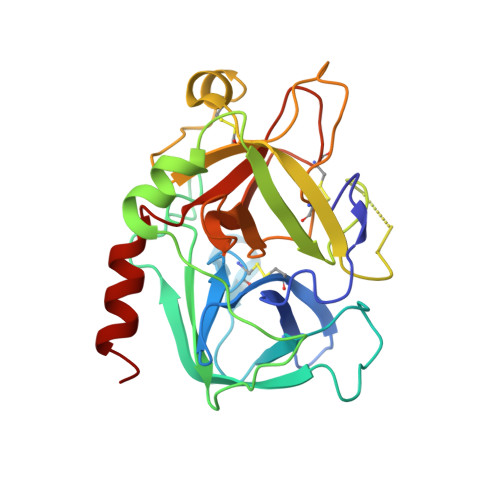
Anopheles mosquitoes are vectors of malaria, a potentially fatal blood disease affecting half a billion humans worldwide. These blood-feeding insects include in their antihemostatic arsenal a potent thrombin inhibitor, the flexible and cysteine-less anophelin. Here, we present a thorough structure-and-function analysis of thrombin inhibition by anophelin, including the 2.3-Å crystal structure of the human thrombin·anophelin complex. Anophelin residues 32-61 are well-defined by electron density, completely occupying the long cleft between the active site and exosite I. However, in striking contrast to substrates, the D50-R53 anophelin tetrapeptide occupies the active site cleft of the enzyme, whereas the upstream residues A35-P45 shield the regulatory exosite I, defining a unique reverse-binding mode of an inhibitor to the target proteinase. The extensive interactions established, the disruption of thrombin's active site charge-relay system, and the insertion of residue R53 into the proteinase S(1) pocket in an orientation opposed to productive substrates explain anophelin's remarkable specificity and resistance to proteolysis by thrombin. Complementary biophysical and functional characterization of point mutants and truncated versions of anophelin unambiguously establish the molecular mechanism of action of this family of serine proteinase inhibitors (I77). These findings have implications for the design of novel antithrombotics.
- Instituto de Biologia Molecular e Celular (IBMC), Universidade do Porto, 4150-180 Porto, Portugal.
Organizational Affiliation: