The Physiological Target for Leurs Translational Quality Control is Norvaline
Cvetesic, N., Palencia, A., Halasz, I., Cusack, S., Gruic-Sovulj, I.(2014) EMBO J 33: 1639
- PubMed: 24935946
- DOI: https://doi.org/10.15252/embj.201488199
- Primary Citation of Related Structures:
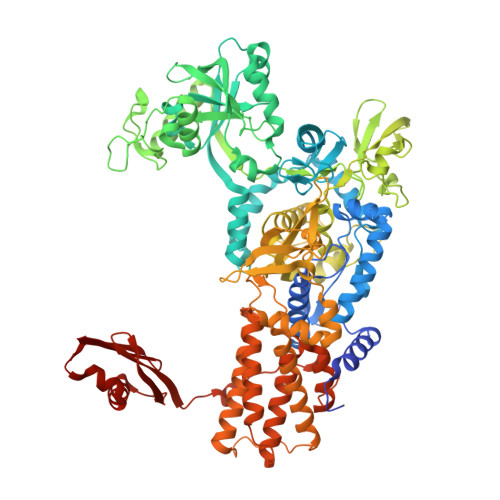
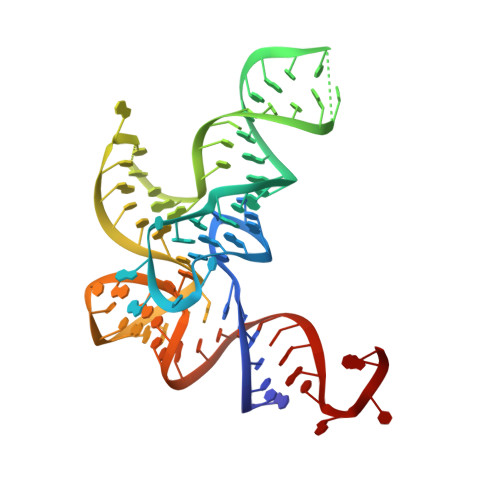
4CQN - PubMed Abstract:
The fidelity of protein synthesis depends on the capacity of aminoacyl-tRNA synthetases (AARSs) to couple only cognate amino acid-tRNA pairs. If amino acid selectivity is compromised, fidelity can be ensured by an inherent AARS editing activity that hydrolyses mischarged tRNAs. Here, we show that the editing activity of Escherichia coli leucyl-tRNA synthetase (EcLeuRS) is not required to prevent incorrect isoleucine incorporation. Rather, as shown by kinetic, structural and in vivo approaches, the prime biological function of LeuRS editing is to prevent mis-incorporation of the non-standard amino acid norvaline. This conclusion follows from a reassessment of the discriminatory power of LeuRS against isoleucine and the demonstration that a LeuRS editing-deficient E. coli strain grows normally in high concentrations of isoleucine but not under oxygen deprivation conditions when norvaline accumulates to substantial levels. Thus, AARS-based translational quality control is a key feature for bacterial adaptive response to oxygen deprivation. The non-essential role for editing under normal bacterial growth has important implications for the development of resistance to antimicrobial agents targeting the LeuRS editing site.
- Department of Chemistry, Faculty of Science University of Zagreb, Zagreb, Croatia.
Organizational Affiliation: