Is the Bovine Lysosomal Phospholipase B-Like Protein an Amidase?
Repo, H., Kuokkanen, E., Oksanen, E., Goldman, A., Heikinheimo, P.(2014) Proteins 82: 300
- PubMed: 23934913
- DOI: https://doi.org/10.1002/prot.24388
- Primary Citation of Related Structures:
4BWC - PubMed Abstract:
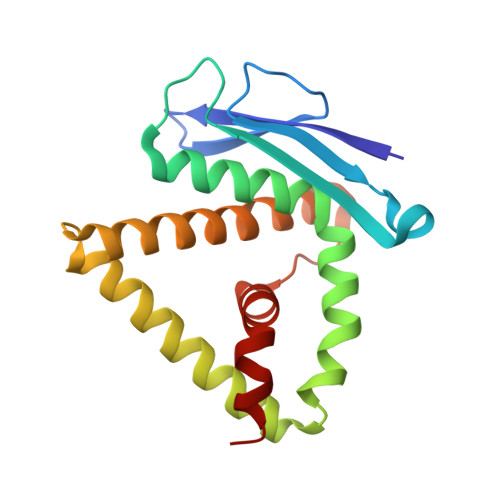
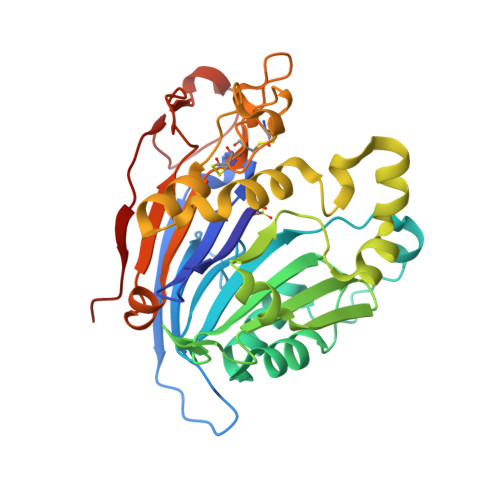
The main function of lysosomal proteins is to degrade cellular macromolecules. We purified a novel lysosomal protein to homogeneity from bovine kidneys. By gene annotation, this protein is defined as a bovine phospholipase B-like protein 1 (bPLBD1) and, to better understand its biological function, we solved its structure at 1.9 Å resolution. We showed that bPLBD1 has uniform noncomplex-type N-glycosylation and that it localized to the lysosome. The first step in lysosomal protein transport, the initiation of mannose-6-phosphorylation by a N-acetylglucosamine-1-phosphotransferase, requires recognition of at least two distinct lysines on the protein surface. We identified candidate lysines by analyzing the structural and sequentially conserved N-glycosylation sites and lysines in bPLBD1 and in the homologous mouse PLBD2. Our model suggests that N408 is the primarily phosphorylated glycan, and K358 a key residue for N-acetylglucosamine-1-phosphotransferase recognition. Two other lysines, K334 and K342, provide the required second site for N-acetylglucosamine-1-phosphotransferase recognition. bPLBD1 is an N-terminal nucleophile (Ntn) hydrolase. By comparison with other Ntn-hydrolases, we conclude that the acyl moiety of PLBD1 substrate must be small to fit the putative binding pocket, whereas the space for the rest of the substrate is a large open cleft. Finally, as all the known substrates of Ntn-hydrolases have amide bonds, we suggest that bPLBD1 may be an amidase or peptidase instead of lipase, explaining the difficulty in finding a good substrate for any members of the PLBD family.
- Institute of Biotechnology, University of Helsinki, FI-00014, Helsinki, Finland; Department of Biosciences, University of Helsinki, FI-00014, Helsinki, Finland.
Organizational Affiliation: