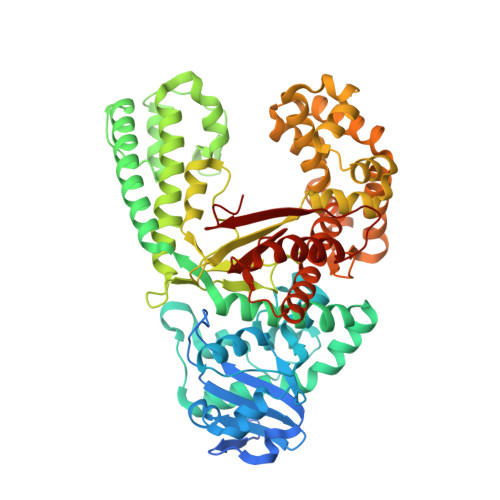
Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Gehrke, T.H., Lischke, U., Gasteiger, K.L., Schneider, S., Arnold, S., Muller, H.C., Stephenson, D.S., Zipse, H., Carell, T.(2013) Nat Chem Biol 9: 455
- PubMed: 23685671
- DOI: https://doi.org/10.1038/nchembio.1254
- Primary Citation of Related Structures:
4B9L, 4B9M, 4B9N, 4B9S, 4B9T, 4B9U, 4B9V - PubMed Abstract:
8-Oxopurines (8-oxodG and 8-oxodA) and formamidopyrimidines (FaPydG and FaPydA) are major oxidative DNA lesions involved in cancer development and aging. Their mutagenicity is believed to result from a conformational shift of the N9-C1' glycosidic bonds from anti to syn, which allows the lesions to form noncanonical Hoogsteen-type base pairs with incoming triphosphates during DNA replication. Here we present biochemical data and what are to our knowledge the first crystal structures of carbocyclic FaPydA and FaPydG containing DNA in complex with a high-fidelity polymerase. Crystallographic snapshots show that the cFaPy lesions keep the anti geometry of the glycosidic bond during error-free and error-prone replication. The observed dG·dC→dT·dA transversion mutations are the result of base shifting and tautomerization.
- Center for Integrated Protein Science at the Department of Chemistry, Ludwig Maximilians University, Munich, Germany.
Organizational Affiliation: