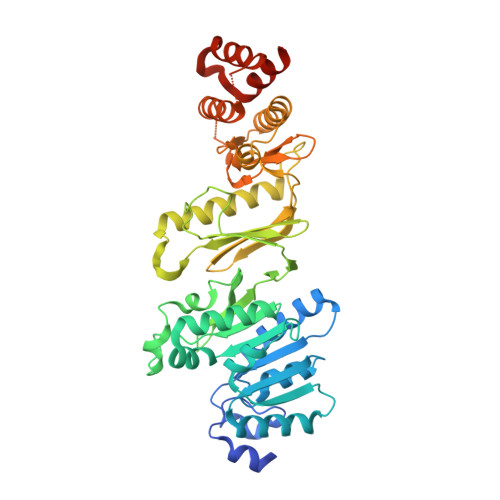
Structure of Wbdd; a Bifunctional Kinase and Methyltransferase that Regulates the Chain Length of the O Antigen in Escherichia Coli O9A.
Hagelueken, G., Huang, H., Clarke, B.R., Lebl, T., Whitfield, C., Naismith, J.H.(2012) Mol Microbiol 86: 730
- PubMed: 22970759
- DOI: https://doi.org/10.1111/mmi.12014
- Primary Citation of Related Structures:
4AZS, 4AZT, 4AZV, 4AZW - PubMed Abstract:
The Escherichia coli serotype O9a O-antigen polysaccharide (O-PS) is a model for glycan biosynthesis and export by the ATP-binding cassette transporter-dependent pathway. The polymannose O9a O-PS is synthesized as a polyprenol-linked glycan by mannosyltransferase enzymes located at the cytoplasmic membrane. The chain length of the O9a O-PS is tightly regulated by the WbdD enzyme. WbdD first phosphorylates the terminal non-reducing mannose of the O-PS and then methylates the phosphate, stopping polymerization. The 2.2 Å resolution structure of WbdD reveals a bacterial methyltransferase domain joined to a eukaryotic kinase domain. The kinase domain is again fused to an extended C-terminal coiled-coil domain reminiscent of eukaryotic DMPK (Myotonic Dystrophy Protein Kinase) family kinases such as Rho-associated protein kinase (ROCK). WbdD phosphorylates 2-α-d-mannosyl-d-mannose (2α-MB), a short mimic of the O9a polymer. Mutagenesis identifies those residues important in catalysis and substrate recognition and the in vivo phenotypes of these mutants are used to dissect the termination reaction. We have determined the structures of co-complexes of WbdD with two known eukaryotic protein kinase inhibitors. Although these are potent inhibitors in vitro, they do not show any in vivo activity. The structures reveal new insight into O-PS chain-length regulation in this important model system.
- Biomedical Sciences Research Complex, University of St Andrews, North Haugh, St Andrews, Fife, KY16 9ST, UK.
Organizational Affiliation: