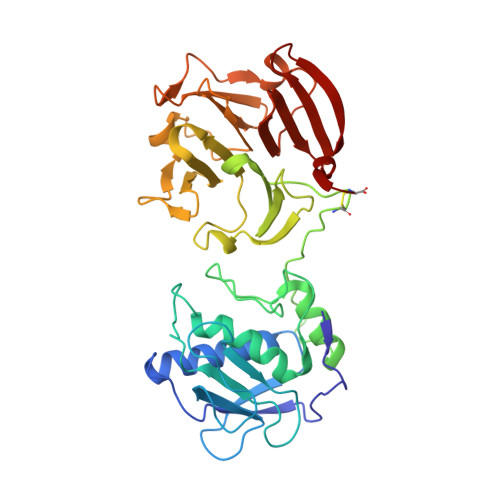
Structural Insights Into Triple-Helical Collagen Cleavage by Matrix Metalloproteinase 1
Manka, S.W., Carafoli, F., Visse, R., Bihan, D., Raynal, N., Farndale, R.W., Murphy, G., Enghild, J.J., Hohenester, E., Nagase, H.(2012) Proc Natl Acad Sci U S A 109: 12461
- PubMed: 22761315
- DOI: https://doi.org/10.1073/pnas.1204991109
- Primary Citation of Related Structures:
4AUO - PubMed Abstract:
Collagenases of the matrix metalloproteinase (MMP) family play major roles in morphogenesis, tissue repair, and human diseases, but how they recognize and cleave the collagen triple helix is not fully understood. Here, we report temperature-dependent binding of a catalytically inactive MMP-1 mutant (E200A) to collagen through the cooperative action of its catalytic and hemopexin domains. Contact between the two molecules was mapped by screening the Collagen Toolkit peptide library and by hydrogen/deuterium exchange. The crystal structure of MMP-1(E200A) bound to a triple-helical collagen peptide revealed extensive interactions of the 115-Å-long triple helix with both MMP-1 domains. An exosite in the hemopexin domain, which binds the leucine 10 residues C-terminal to the scissile bond, is critical for collagenolysis and represents a unique target for inhibitor development. The scissile bond is not correctly positioned for hydrolysis in the crystallized complex. A productive binding mode is readily modeled, without altering the MMP-1 structure or the exosite interactions, by axial rotation of the collagen homotrimer. Interdomain flexing of the enzyme and a localized excursion of the collagen chain closest to the active site, facilitated by thermal loosening of the substrate, may lead to the first transition state of collagenolysis.
- Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences, Kennedy Institute of Rheumatology, University of Oxford, London W6 8LH, United Kingdom.
Organizational Affiliation: