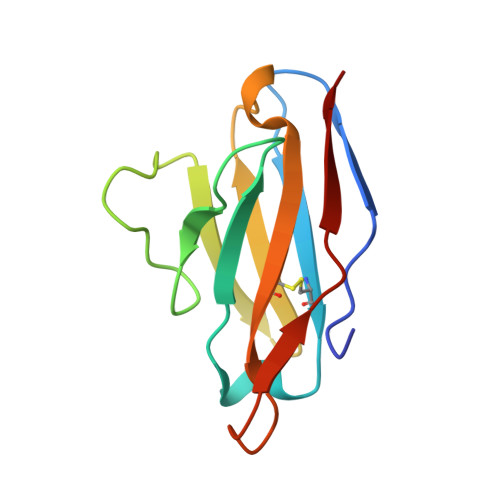
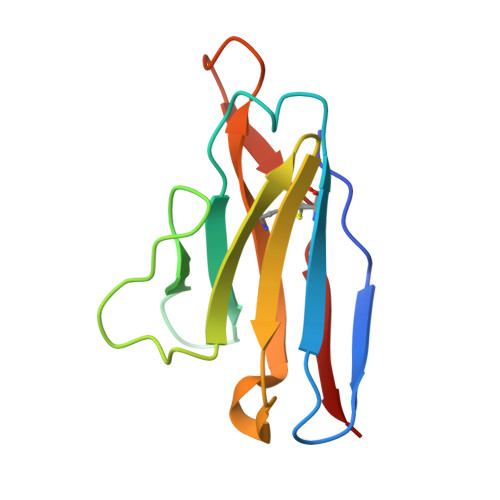
Site-Directed Mutagenesis Reveals Regions Implicated in the Stability and Fiber Formation of Human Lambda3R Light Chains.
Villalba, M.I., Canul-Tec, J.C., Luna-Martinez, O.D., Sanchez-Alcala, R., Olamendi-Portugal, T., Rudino-Pinera, E., Rojas, S., Sanchez-Lopez, R., Fernandez-Velasco, D.A., Becerril, B.(2015) J Biological Chem 290: 2577
- PubMed: 25505244
- DOI: https://doi.org/10.1074/jbc.M114.629550
- Primary Citation of Related Structures:
4AIX, 4AIZ, 4AJ0 - PubMed Abstract:
Light chain amyloidosis (AL) is a disease that affects vital organs by the fibrillar aggregation of monoclonal light chains. λ3r germ line is significantly implicated in this disease. In this work, we contrasted the thermodynamic stability and aggregation propensity of 3mJL2 (nonamyloidogenic) and 3rJL2 (amyloidogenic) λ3 germ lines. Because of an inherent limitation (extremely low expression), Cys at position 34 of the 3r germ line was replaced by Tyr reaching a good expression yield. A second substitution (W91A) was introduced in 3r to obtain a better template to incorporate additional mutations. Although the single mutant (C34Y) was not fibrillogenic, the second mutation located at CDR3 (W91A) induced fibrillogenesis. We propose, for the first time, that CDR3 (position 91) affects the stability and fiber formation of human λ3r light chains. Using the double mutant (3rJL2/YA) as template, other variants were constructed to evaluate the importance of those substitutions into the stability and aggregation propensity of λ3 light chains. A change in position 7 (P7D) boosted 3rJL2/YA fibrillogenic properties. Modification of position 48 (I48M) partially reverted 3rJL2/YA fibril aggregation. Finally, changes at positions 8 (P8S) or 40 (P40S) completely reverted fibril formation. These results confirm the influential roles of N-terminal region (positions 7 and 8) and the loop 40-60 (positions 40 and 48) on AL. X-ray crystallography revealed that the three-dimensional topology of the single and double λ3r mutants was not significantly altered. This mutagenic approach helped to identify key regions implicated in λ3 AL.
- From the Departments of Molecular Medicine and Bioprocesses and.
Organizational Affiliation: