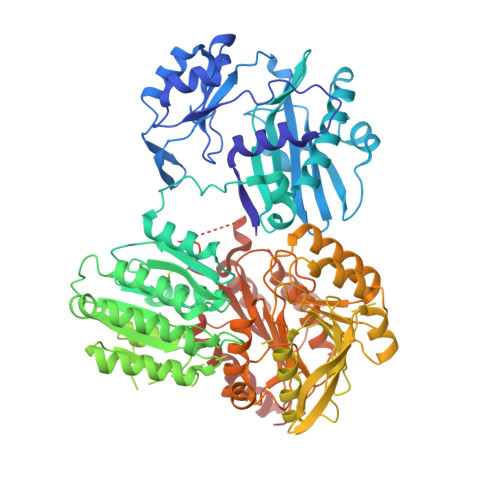
ADP-Mg2+ bound to the ATP-grasp domain of ATP-citrate lyase.
Sun, T., Hayakawa, K., Fraser, M.E.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 1168-1172
- PubMed: 22102020
- DOI: https://doi.org/10.1107/S1744309111028363
- Primary Citation of Related Structures:
3PFF - PubMed Abstract:
Human ATP-citrate lyase (EC 2.3.3.8) is the cytoplasmic enzyme that catalyzes the production of acetyl-CoA from citrate, CoA and ATP. The amino-terminal portion of the enzyme, containing residues 1-817, was crystallized in the presence of tartrate, ATP and magnesium ions. The crystals diffracted to 2.3 Å resolution. The structure shows ADP-Mg(2+) bound to the domain that possesses the ATP-grasp fold. The structure demonstrates that this crystal form could be used to investigate the structures of complexes with inhibitors of ATP-citrate lyase that bind at either the citrate- or ATP-binding site.
- Department of Biological Sciences, University of Calgary, 2500 University Drive NW, Calgary, AB T2N 1N4, Canada.
Organizational Affiliation: