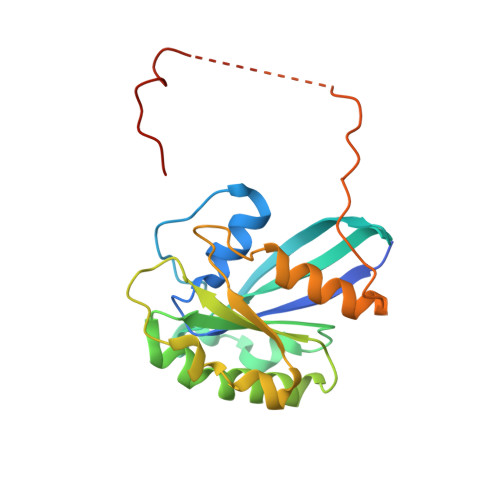
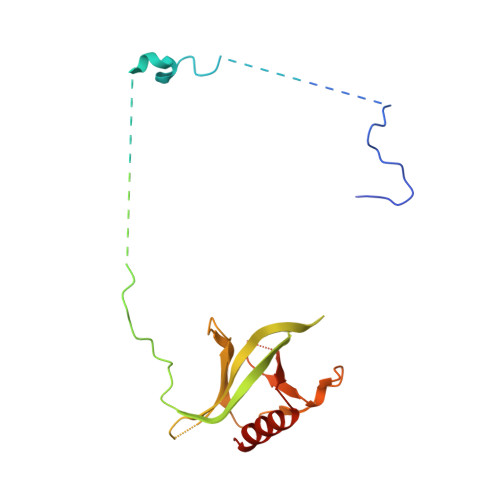
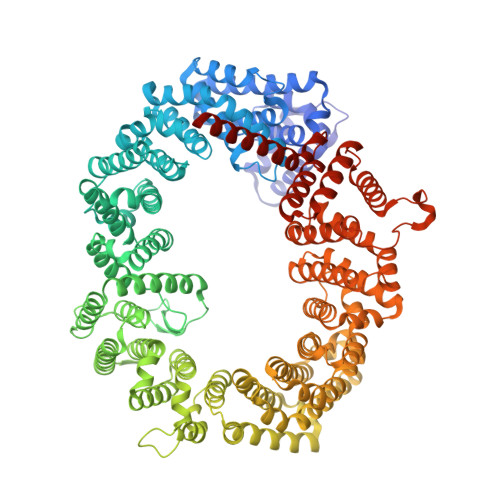
Structural insights into how yrb2p accelerates the assembly of the xpo1p nuclear export complex
Koyama, M., Shirai, N., Matsuura, Y.(2014) Cell Rep 9: 983-995
- PubMed: 25437554
- DOI: https://doi.org/10.1016/j.celrep.2014.09.052
- Primary Citation of Related Structures:
3WYF, 3WYG - PubMed Abstract:
Proteins and ribonucleoproteins containing a nuclear export signal (NES) assemble with the exportin Xpo1p (yeast CRM1) and Gsp1p-GTP (yeast Ran-GTP) in the nucleus and exit through the nuclear pore complex. In the cytoplasm, Yrb1p (yeast RanBP1) displaces NES from Xpo1p. Efficient export of NES-cargoes requires Yrb2p (yeast RanBP3), a primarily nuclear protein containing nucleoporin-like phenylalanine-glycine (FG) repeats and a low-affinity Gsp1p-binding domain (RanBD). Here, we show that Yrb2p strikingly accelerates the association of Gsp1p-GTP and NES to Xpo1p. We have solved the crystal structure of the Xpo1p-Yrb2p-Gsp1p-GTP complex, a key assembly intermediate that can bind cargo rapidly. Although the NES-binding cleft of Xpo1p is closed in this intermediate, our data suggest that preloading of Gsp1p-GTP onto Xpo1p by Yrb2p, conformational flexibility of Xpo1p, and the low affinity of RanBD enable active displacement of Yrb2p RanBD by NES to occur effectively. The structure also reveals the major binding sites for FG repeats on Xpo1p.
- Division of Biological Science, Graduate School of Science, Nagoya University, 464-8602 Furo-cho, Chikusa-ku, Nagoya City, Japan.
Organizational Affiliation: