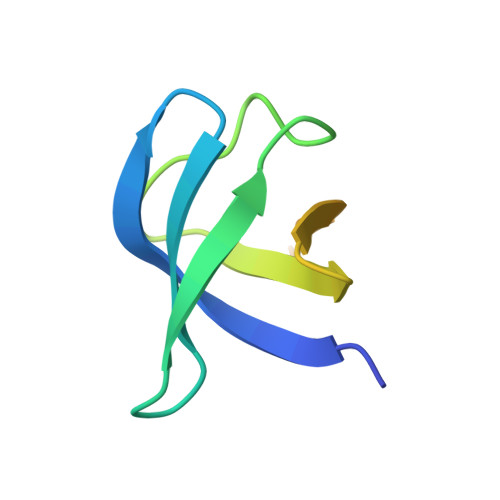
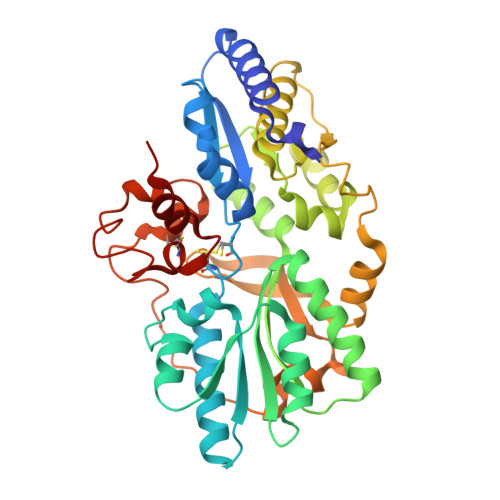
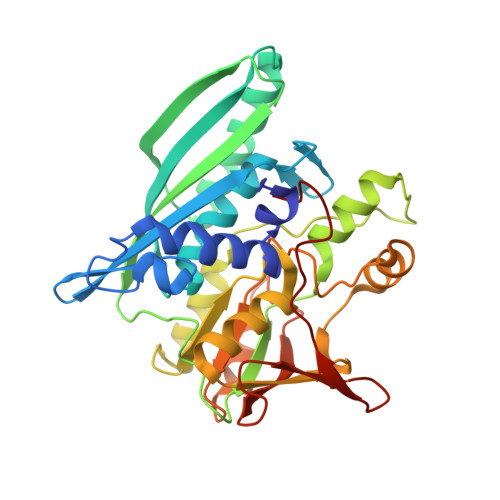
Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Watanabe, S., Matsumi, R., Atomi, H., Imanaka, T., Miki, K.(2012) Structure 20: 2124-2137
- PubMed: 23123111
- DOI: https://doi.org/10.1016/j.str.2012.09.018
- Primary Citation of Related Structures:
3VYR, 3VYS, 3VYT, 3VYU - PubMed Abstract:
[NiFe] hydrogenase maturation represents one of the most dynamic and sophisticated processes in metallocenter assembly. The Fe(CN)(2)CO moiety of [NiFe] hydrogenases is assembled via unknown transient interactions among specific maturation proteins HypC (metallochaperone), HypD (redox protein), and HypE (cyanide synthesis/donor). Here, we report the structures of the HypC-HypD and HypC-HypD-HypE complexes, providing a view of the transient interactions that take place during the maturation process. HypC binds to the conserved region of HypD through extensive hydrophobic interactions. The ternary complex formation between HypE and the HypCD complex involves both HypC and HypD, rendering the HypE conformation favorable for cyanide transfer. In the complex, the conserved cysteines of HypC and HypD form an Fe binding site. The conserved C-terminal cysteine of HypE can access the thiol redox cascade of HypD. These results provide structural insights into the Fe atom cyanation in the HypCDE complex.
- Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan.
Organizational Affiliation: