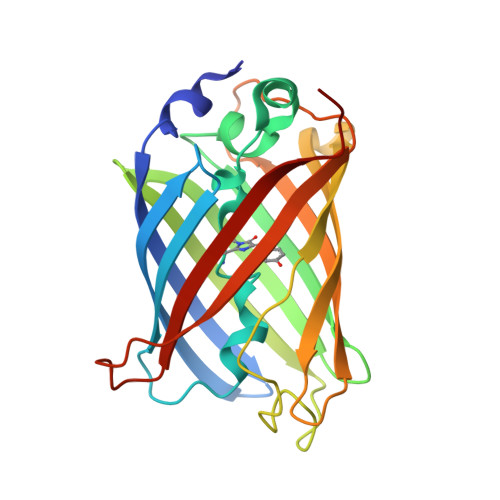
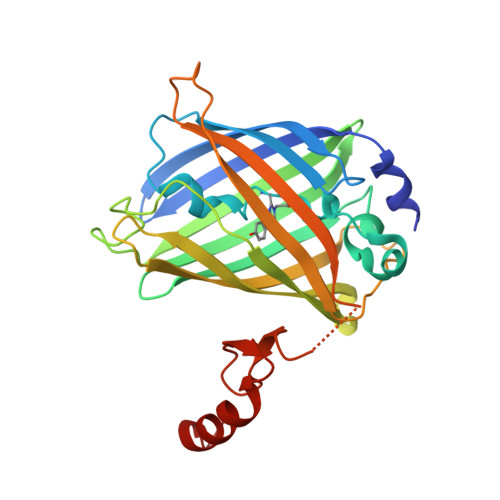
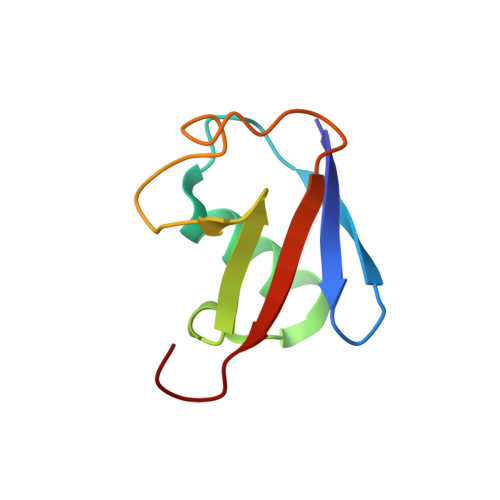
A novel mode of ubiquitin recognition by the ubiquitin-binding zinc finger domain of WRNIP1.
Suzuki, N., Rohaim, A., Kato, R., Dikic, I., Wakatsuki, S., Kawasaki, M.(2016) FEBS J
- PubMed: 27062441
- DOI: https://doi.org/10.1111/febs.13734
- Primary Citation of Related Structures:
3VHS, 3VHT, 3WUP, 4Z4K, 4Z4M - PubMed Abstract:
The ubiquitin-binding zinc finger (UBZ) is a type of zinc-coordinating β-β-α fold domain found mainly in proteins involved in DNA repair and transcriptional regulation. Here, we report the crystal structure of the UBZ domain of Y-family DNA polymerase (pol) η and the crystal structure of the complex between the UBZ domain of Werner helicase-interacting protein 1 (WRNIP1) and ubiquitin, crystallized using the GFP fusion technique. In contrast to the pol η UBZ, which has been proposed to bind ubiquitin via its C-terminal α-helix, ubiquitin binds to a novel surface of WRNIP1 UBZ composed of the first β-strand and the C-terminal α-helix. In addition, we report the structure of the tandem UBZ domains of Tax1-binding protein 1 (TAX1BP1) and show that the second UBZ of TAX1BP1 binds ubiquitin, presumably in a manner similar to that of WRNIP1 UBZ. We propose that UBZ domains can be divided into at least two different types in terms of the ubiquitin-binding surfaces: the pol η type and the WRNIP1 type. Structural data are available in the Protein Data Bank under accession numbers 3WUP (pol η UBZ), 3VHS (WRNIP1 UBZ), 3VHT (GFP-WRNIP1/ubiquitin), 4Z4K (TAX1BP1 UBZ1 + 2), and 4Z4M (TAX1BP1 UBZ2).
- Structural Biology Research Center, Photon Factory, Institute of Materials Structure Science, High Energy Accelerator Research Organization (KEK), Tsukuba, Ibaraki, Japan.
Organizational Affiliation: