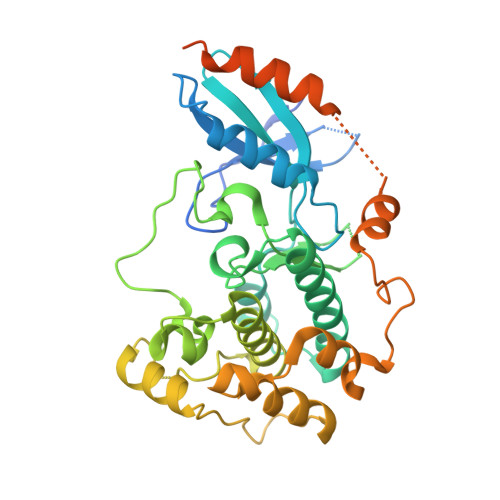
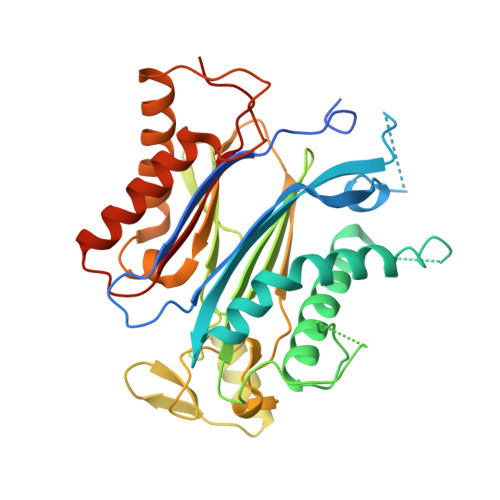
Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases.
Soon, F.F., Ng, L.M., Zhou, X.E., West, G.M., Kovach, A., Tan, M.H., Suino-Powell, K.M., He, Y., Xu, Y., Chalmers, M.J., Brunzelle, J.S., Zhang, H., Yang, H., Jiang, H., Li, J., Yong, E.L., Cutler, S., Zhu, J.K., Griffin, P.R., Melcher, K., Xu, H.E.(2012) Science 335: 85-88
- PubMed: 22116026
- DOI: https://doi.org/10.1126/science.1215106
- Primary Citation of Related Structures:
3UJG, 3UJK, 3UJL - PubMed Abstract:
Abscisic acid (ABA) is an essential hormone for plants to survive environmental stresses. At the center of the ABA signaling network is a subfamily of type 2C protein phosphatases (PP2Cs), which form exclusive interactions with ABA receptors and subfamily 2 Snfl-related kinase (SnRK2s). Here, we report a SnRK2-PP2C complex structure, which reveals marked similarity in PP2C recognition by SnRK2 and ABA receptors. In the complex, the kinase activation loop docks into the active site of PP2C, while the conserved ABA-sensing tryptophan of PP2C inserts into the kinase catalytic cleft, thus mimicking receptor-PP2C interactions. These structural results provide a simple mechanism that directly couples ABA binding to SnRK2 kinase activation and highlight a new paradigm of kinase-phosphatase regulation through mutual packing of their catalytic sites.
- Laboratory of Structural Sciences, Van Andel Research Institute, 333 Bostwick Avenue NE, Grand Rapids, MI 49503, USA.
Organizational Affiliation: