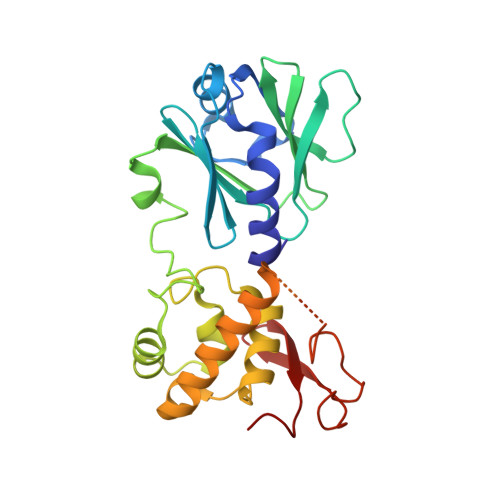
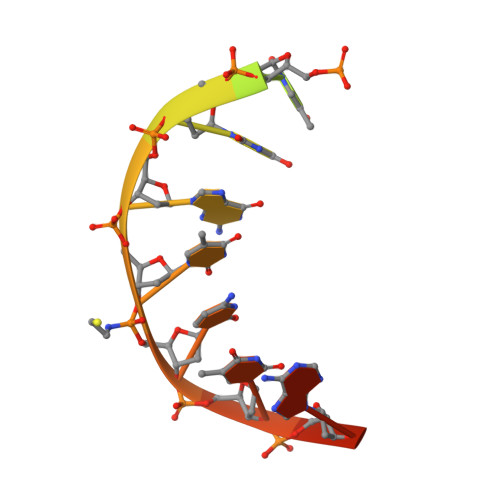
Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
Sung, R.J., Zhang, M., Qi, Y., Verdine, G.L.(2012) J Biological Chem 287: 18044-18054
- PubMed: 22465958
- DOI: https://doi.org/10.1074/jbc.M111.313635
- Primary Citation of Related Structures:
3U6C, 3U6D, 3U6E, 3U6L, 3U6M, 3U6O, 3U6P, 3U6Q, 3U6S - PubMed Abstract:
MutM, a bacterial DNA-glycosylase, plays a critical role in maintaining genome integrity by catalyzing glycosidic bond cleavage of 8-oxoguanine (oxoG) lesions to initiate base excision DNA repair. The task faced by MutM of locating rare oxoG residues embedded in an overwhelming excess of undamaged bases is especially challenging given the close structural similarity between oxoG and its normal progenitor, guanine (G). MutM actively interrogates the DNA to detect the presence of an intrahelical, fully base-paired oxoG, whereupon the enzyme promotes extrusion of the target nucleobase from the DNA duplex and insertion into the extrahelical active site. Recent structural studies have begun to provide the first glimpse into the protein-DNA interactions that enable MutM to distinguish an intrahelical oxoG from G; however, these initial studies left open the important question of how MutM can recognize oxoG residues embedded in 16 different neighboring sequence contexts (considering only the 5'- and 3'-neighboring base pairs). In this study we set out to understand the manner and extent to which intrahelical lesion recognition varies as a function of the 5'-neighbor. Here we report a comprehensive, systematic structural analysis of the effect of the 5'-neighboring base pair on recognition of an intrahelical oxoG lesion. These structures reveal that MutM imposes the same extrusion-prone ("extrudogenic") backbone conformation on the oxoG lesion irrespective of its 5'-neighbor while leaving the rest of the DNA relatively free to adjust to the particular demands of individual sequences.
- Department of Molecular and Cellular Biology, Harvard University, Cambridge, Massachusetts 02138, USA.
Organizational Affiliation: